疟疾是由疟原虫引起的一种全球性疾病。据世界卫生组织(WHO)统计,2015年有2.14亿例疟疾发生,并有43.8万人死于疟疾[1]。尽管在控制疟疾方面已经做了极大努力,但疟疾在全球的发病率和死亡率仍然很高。以青蒿素为基础的联合用药(ACTs)在治疗疟疾方面疗效显著,成为抗疟一线药物[2]。
青蒿素(artemisinin)是一种含过氧桥基团结构的倍半萜内酯类化合物,主要来源于我国传统药用植物黄花蒿Artemisia annua L.(以下简称“青蒿”)。目前,青蒿素市场需求巨大,然而青蒿中青蒿素的量很低,仅占干质量的0.01%~1%[3]。为缓解青蒿素市场供求的巨大矛盾,科学家就提高青蒿素的量进行了大量研究。本文对青蒿素代谢调控的研究进展进行了综述,以期为提高青蒿素量提供新的思路,并为培育优质、高产的青蒿品系奠定基础。
1 青蒿素及相关萜类生物合成途径萜类起源于异戊烯基焦磷酸(isopentenyl diphosphate,IPP)和二甲基丙烯基焦磷酸(dimethylallyl diphosphate,DMAPP),包含甲羟戊酸途径(MVA途径,位于胞质)和磷酸甲基赤藓糖途径(MEP途径,位于质体)。法尼基焦磷酸(farnesyl iphosphate,FPP)是合成多种次生代谢产物的底物,Schramek等[4]通过13CO2同位素标记证明,一分子MEP途径的IPP和一分子MVA途径的DMAPP生成一分子牻牛儿基二磷酸(geranyl diphosphate,GPP),然后GPP进入胞质(cytoplasm),与一分子MEP途径的IPP结合生成一分子FPP。如图 1所示,FPP是多种萜类化合物生物合成的关键中间体。
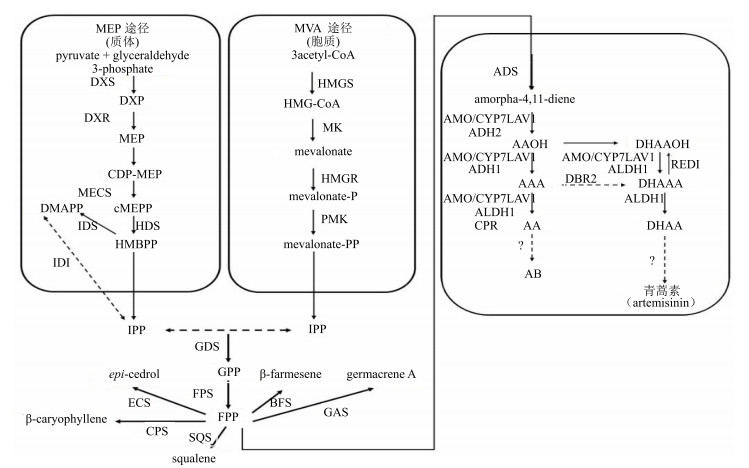
|
ADS-紫穗槐二烯合成酶ALDH1-醛脱氢酶1 AMO/CYP71AV1-紫穗槐-氧化酶AAOH-青蒿醇AAA-青蒿醛AA-青蒿酸AB-青蒿素B ECS-雪松醇合酶SQS-角鲨烯合酶GAS-大根香叶烯A合成酶BFS-β-法尼烯合成酶CPS-β-石竹烯合成酶CPR-细胞色素P450还原酶RED1-二氢青蒿醛还原酶CYP71AV1-细胞色素P450单氧化酶DBR2-青蒿醛双键还原酶ADH1-乙醇脱氢酶1 ADH2-乙醇脱氢酶2 DHAAOH-双氢青蒿醇DHAAA-二氢青蒿醛DHAA-二氢青蒿酸DMAPP-二甲基烯丙基焦磷酸酯FPS-法尼基焦磷酸合成酶FPP-法尼基焦磷酸GPP-牻牛儿基二磷酸HMGR-3-羟基-3-甲基-戊二酰辅酶A还原酶HMGS-3-羟基-3-甲基-戊二酰辅酶A合成酶IPP-异戊烯焦磷酸IDS-IPP/DMAPP合成酶MVA-甲羟戊酸CDP-MEP-二磷酸胞苷-2-C-甲基-D-赤藻糖醇-2-磷酸cMEPP-2-甲基-D-赤藻糖醇-2, 4-环二磷酸酯DXP-1-脱氧-D-木酮糖-5-磷酸DXR-1-脱氧-D-木酮糖-5-磷酸还原异构化酶DXS-1-脱氧木酮糖-5-磷酸合成酶GDS-牻牛儿基二磷酸合成酶HDS-4-羟基-2-甲基-2-E-丁烯基-4-焦磷酸合酶HMBPP-1-羟基-2-甲基-2-丁烯-4-焦磷酸MECS-2-C-甲基-D-赤藓糖-2, 4-环焦磷酸合酶MEP-2-甲基-D-赤藻糖醇-4-磷酸MK-甲羟戊酸激酶PMK-二氧磷基甲羟戊酸激酶
ADS-amorpha-4, 11-diene synthase ALDH1-aldehyde dehydrogenase 1 AMO/CYP71AV1-amorphadiene-12-hydroxylase AAOH-artemisinic alcohol AAA-artemisinic aldehyde AA-artemisinic acid AB-arteannuin B ECS-epi-cedrol synthase SQS-squalene synthase GAS-germacrene synthase A BFS- |
在青蒿素生物合成途径中,第一步特异性反应是由FPP生成紫穗槐二烯(amorpha-4, 11-diene)。MVA和MEP途径共同提供青蒿素生物合成所需的IPP,2分子IPP结合DMAPP生成GPP,最后形成FPP。FPP经紫穗槐二烯合成酶(amorpha-4, 11-diene synthase,ADS)催化,生成紫穗槐二烯[5];紫穗槐二烯经由细胞色素P450单氧化酶(cytochrome P450 monooxygenase,CYP71AV1)催化的3步反应,分别形成青蒿醇(artemisinic alcohol,AAOH)、青蒿醛(artemisinic aldehyde,AAA)和青蒿酸(artemisinic acid,AA)[6-8]。其中,AAOH可以被催化形成二氢青蒿醇(dihydroartemisinic alcohol,DHAAOH)[9];AAA可以被青蒿醛双键还原酶[artemisinic aldehyde delta-11(13) reductase,DBR2]催化形成二氢青蒿醛(dihydroartemisinic aldehyde,DHAAA)[9];AA可以生成终产物青蒿素,也可经青蒿烯形成终产物青蒿素B[10-11]。另外,DHAAOH被CYP71AV1和ALDH1催化形成DHAAA,后经二氢青蒿酸(dihydroartemisinic acid,DHAA)形成青蒿素[11];DHAAA一方面可被醛脱氢酶1(aldehyde dehydrogenase 1,ALDH1)催化形成DHAA[12],另一方面也能由二氢青蒿醛还原酶(dihydroartemisinic aldehyde reductase,RED1)还原形成DHAAOH[13]。虽然DHAA已经被证实为青蒿素的直接前体,但这一转化在生物体内是否为酶促反应仍然存在争议[11]。
除青蒿素生物合成途径外,FPP经由不同的酶催化可以形成其他4个倍半萜和1个三萜。以FPP为底物,经雪松醇合酶(epi-cedrol synthase,ECS)可催化生成β-雪松醇倍半萜[14];经另一个关键酶β-石竹烯合酶(β-caryophyllene synthase,CPS),可催化FPP生成β-石竹烯倍半萜[15];而经大根香叶烯合酶(germacrene A synthase,GAS)基因编码的酶能催化FPP合成大根香叶烯A倍半萜[16];另外,FPP经β-法尼烯合成酶(β-farnesene synthase,BFS)能催化产生单一的β-法尼烯倍半萜[17-18];除此之外,研究已经证明FPP还可被角鲨烯合酶(squalene synthesisase,SQS)催化形成角鲨烯三萜或甾体[19-20]。
2 青蒿素相关合成途径关键酶基因对青蒿素合成的影响随着分子生物学技术的发展,已经克隆到众多青蒿素及其相关合成途径的关键酶基因。通过过量表达青蒿素生物合成途径的关键酶基因及抑制合成代谢支路途径等手段,可在一定程度上增加通往青蒿素生物合成途径的代谢流,提高青蒿中青蒿素的量。基于目前已经报道的青蒿素及相关萜类生物合成途径,本文归纳了所有可以进行代谢调控的基因,结果见表 1。其中,已进行单基因过表达的青蒿素生物合成途经相关基因有HMGR、FPS、DXR、DBR2、ALDH1和ADS;已进行多基因过表达的基因组合形式有HDR/ ADS[28]、FPS/ADS[29]、CYP71AV1/CPR、HMGR/ ADS、HMGR/FPS、ADS/CYP71AV1/CPR和FPS/CYP71AV1/CPR等[29]。也有研究证明抑制青蒿素合成的竞争支路途径基因SQS、CPS、BFS、GAS等,也能提高青蒿素的量[27]。
|
|
表 1 青蒿素合成途径及支路途径关键酶基因 Table 1 Genes of artemisinin synthesis pathway and branch pathway |
早期研究已经证实,青蒿素由分泌型腺毛产生[30-32]。目前所报道的青蒿素合成相关的基因都在腺毛中高表达或特异性表达[33];此外,一些腺毛相关或特异性表达的基因通过调控腺毛或青蒿素生物合成相关基因,继而影响青蒿素的合成[34];腺毛还具备青蒿素生物合成所必需的特殊油性氧化环境[35]。研究表明,分泌型腺毛密度与青蒿素量呈正相关[5, 36-37];糖苷水解酶(β-glucosidase)转基因青蒿可通过调控腺毛密度增加青蒿素的量[38]。
DHAA是青蒿素合成的直接前体,其合成后的转运对于青蒿素的合成十分关键。近期的研究结果表明,脂质转运蛋白3(AaLTP3)和多向耐药性转运蛋白(AaPDR2)能提高(DH)AA在烟草叶质外体中的积累量,防止(DH)AA从质外体到细胞的逆向流,最终从总体上增强AB和青蒿素的代谢流。这说明转运蛋白也可作为调控青蒿素生物合成的靶标基因[39]。
另外,在青蒿次生代谢调控中,启动子也是一个关键因素。Han等[40]的实验结果表明,CYP71AV1启动子和CaMV35S启动子分别调控FPS/ADS基因组合表达时,CaMV35S启动子驱动的基因的表达量较高,但腺毛特异性启动子CYP71AV1能驱动青蒿素合成途径基因在腺毛中特异表达,更利于青蒿素的合成。该结果表明,在青蒿次生代谢调控中使用青蒿素生物合成关键基因的启动子,如CYP71AV1启动子,更有利于转基因植株中青蒿素的积累。
3 转录因子对青蒿素合成的影响转录因子(transcription factor,TF)是一类能特异结合基因启动子的蛋白质,是植物代谢工程中的一种重要手段。转录因子通过与基因启动子上相应的顺式作用元件结合,从而激活目标基因的转录。目前在青蒿素生物合成过程中发现有多种转录因子参与,包括WRKY(1个)、NAC(1个)、bHLH(2个)、bZIP(2个)和AP2/ERF(4个)转录因子家族。它们通过调控青蒿素生物合成途径基因的表达或青蒿表面腺毛的密度,最终影响青蒿素的合成。
3.1 WRKY家族转录因子WRKY是植物特有的转录因子家族,AaWRKY1是最早在青蒿中发现的转录因子,研究表明该基因可与ADS和CYP71AV1启动子区域的W-box结合,WRKY1过表达可提高转基因植株中HMGR、DBR2、ADS和CYP71AV1基因的表达水平,增加青蒿素的积累[41-42]。
3.2 NAC家族转录因子NAC类转录因子是植物中最大类转录因子之一,该家族的转录因子AaNAC1受水杨酸(salicylic acid,SA)、茉莉酸(jasmonic acid,JA)和干旱诱导,在青蒿中过表达该基因,导致青蒿素合成途径基因ADS的表达量升高,青蒿素量增加[43]。
3.3 bHLH家族转录因子AabHLH1和AaMYC2属于helix loop-helix(bHLH)类转录因子。AabHLH1从青蒿分泌型腺毛的cDNA文库中分离获得,研究证明该基因可与ADS和CYP71AV1启动子区域的E-box顺式作用元件结合,在青蒿中过表达AabHLH1可提高HMGR、ADS和CYP71AV1基因的表达水平[44]。Shen等[45]在青蒿中克隆获得AaMYC2转录因子,证明该基因受JA诱导,可与CYP71AV1和DBR2启动子中的G-box like结构域结合,过表达AaMYC2基因上调了CYP71AV1和DBR2基因的转录水平,增加了青蒿素的量;另有研究表明,在青蒿倍半萜合成中,JA与GA之间的信号传导依赖AaMYC2-DELLA蛋白的相互作用[46]。
3.4 bZIP家族转录因子AabZIP1和AaHD1同属于bZIP类转录因子家族,具有基本的DNA结合区域和亮氨酸拉链结构。研究证明,AabZIP1基因通过与ADS和CYP71AV1启动子区域的ABA反应元件(ABRE)结合,上调了ADS、CYP71AV1、DBR2和ALDH1基因的转录水平,影响青蒿素的生物合成[47];AabZIP1将ABA信号通路与青蒿素生物合成联系起来。而AaHD1与JA信号传导相关,该基因通过结合AaJAZ8(jasmonate ZIM-domain 8),影响JA活性,调控青蒿中腺毛的形成,AaHD1过表达可显著增加青蒿分泌型腺毛的密度,最终提高青蒿中青蒿素的量[48]。
3.5 AP2/ERF家族转录因子AP2/ERF转录因子家族在青蒿转录调控中研究较多,目前已经报道的AaERF1、AaERF2、AaORA和AaTAR1均属于该家族。Yu等[49]研究表明AaERF1和AaERF2为乙烯和JA响应的AP2转录因子,与ADS和CYP71AV1启动子区域的CRTDREHVCBF2(CBF2)和RAV1AAT(RAA)结合,调控ADS和CYP71AV1基因的表达,提高了青蒿素的量。Lu等[50]通过实验证明,AaORA可调控腺毛形成,通过防御标志基因PDF1.2、HEL和B-CHI影响青蒿对灰霉菌Botryris cinerea的抵抗作用,正向调节DBR2、ADS、CYP71AV1和AaERF1的转录水平,增加青蒿素的量。本课题组克隆了另一个AP2/ERF转录因子--AaTAR1,证明该基因可影响青蒿腺毛发育及蜡质合成,还可通过与ADS和CYP71AV1启动子区域的CBF2和RAA结合,调控青蒿素的生物合成[51]。另外,Wang等[52]通过对ALDH1启动子的克隆,证明AaORA和AaERF2也可与RAA结构域相互作用,影响青蒿素的合成。
转录因子种类繁多,除以上已经报道的转录因子家族外,最近还报道了2个在青蒿腺毛中特异性表达的其他家族转录因子AaGL2和AaMIXTA-Like1[53]。转录因子在植物次生代谢调控中具有不可替代的作用,目前已经报道的在青蒿中表达的转录因子有10个左右,一些重要的更有潜力的转录因子可发掘作为青蒿代谢调控的靶标基因。
4 植物激素对青蒿素合成的影响植物激素是一种信号分子,可以调节特定的细胞进程,对于植物生长发育具有十分重要的作用。植物内源性信号分子JA、赤霉素(gibberellins,GA)、脱落酸(abscisic acid,ABA)和SA可通过转录因子调控青蒿素生物合成和腺毛的发育,影响青蒿素的合成。
4.1 JA对青蒿素合成的影响JA属于脂类衍生物,为保守的植物次生代谢激发因子[54-55]。青蒿中已经报道的多种转录因子均受JA诱导,包括AaNAC1[43]、AaMYC2[45]、AaERF1和AaERF2[49]等。这些转录因子经JA诱导后表达量增加,从而正向调控青蒿素生物合成途径中的多个关键酶基因,提高青蒿素的量。此外,JA还可影响青蒿腺毛密度。由于青蒿素和腺毛密度密切相关[56-57],JA可通过影响腺毛相关基因,如TFAR1(trichome-specific fatty acyl-CoA reductase 1),调控腺毛数量[57];JA还可借助bZIP类转录因子家族的AaHD1诱导腺毛的形成[48]。
4.2 ABA对青蒿素合成的影响ABA是一种倍半萜烯类植物激素,在非生物胁迫应答中起重要作用[58]。研究表明,用ABA处理青蒿悬浮细胞[59],可使青蒿素合成途径关键基因HMGR、FPS、CYP71AV1的表达量显著升高,继而增加青蒿素产量。Zhang等[60]在青蒿中克隆出了ABA受体AaPYL9,并证明在青蒿中过表达AaPYL9能提高青蒿素合成途径基因FPS、ADS和CYP71AV1的转录水平,增加青蒿中青蒿素和二氢青蒿酸的量。Zhang等[47]又在青蒿中克隆获得ABA途径的ABF类转录因子--AabZIP1,该基因能直接结合青蒿素合成途径基因ADS和CYP71AV1的启动子,提高青蒿素的生物合成能力。
4.3 SA对青蒿素合成的影响SA是一种酚类化合物,也能影响植物中次生代谢产物的合成。研究表明,向青蒿植株喷洒外源SA时,青蒿素生物合成途径关键基因ADS的表达量增加,青蒿素和二氢青蒿酸的量增加[61-62]。研究表明,SA在青蒿中可能通过2种途径增加青蒿素的量:一是提高活性氧(ROS)的量,促进二氢青蒿酸向青蒿素转化;二是影响青蒿素生物合成相关的酶基因[1]。但SA提高青蒿素的分子机制还有待进一步研究。
4.4 GA对青蒿素合成的影响GA属于二萜类酸,是目前发现能促进青蒿素生物合成的最重要的植物激素[1]。内源性GA可诱导AA向青蒿素转化[63];GA能提高青蒿素生物合成途径中FDS、ADS和CYP71AV1基因的表达量[57, 64]。早期的研究也表明,当用内源性GA处理青蒿后,青蒿素的量增加了3~4倍,同时腺毛数量增加[65]。但目前基于GA调控青蒿素生物合成及腺毛的分子机制的研究还未见报道。
JA、GA、ABA和SA这几种植物激素虽然有其特定的生物合成途径及信号传导通路,但它们之间也存在相互作用网络。JA与GA之间可依赖AaMYC2-DELLA基因的相互作用进行信号传导[46];GA诱导DELLA蛋白生长抑制因子降解,提高ROS和SA的积累,削弱JA信号通路[66];JA与SA信号相互拮抗[56-67]。
5 逆境及诱导因子对青蒿素合成的影响 5.1 逆境对青蒿素合成的影响各种环境胁迫因子可作为增加植物次生代谢产物的诱导因子。当植物在细胞水平上识别刺激信号后,即产生应激反应。环境胁迫信号如温度(高、低),高盐,水(干旱、洪灾),射线,化学物质,金属及生物胁迫通常会诱导苯丙烷类物质的累积[67]。
将青蒿暴露在不同的诱导因子下,研究青蒿素的产量及其生物合成途径相关基因转录水平的变化。研究[67]显示,寒冷可以增加青蒿素的产量,主要是通过增加青蒿素合成途径基因ADS、CYP71AV1和DXS的表达量;夜雾和干旱也能提高青蒿素的产量。高盐对青蒿素产量的影响具有两面性,在植物生长早期主要通过增加氧化应激促进AA向青蒿素的转化,提高青蒿素的量;在植物的生长后期,高盐却抑制青蒿素的合成。另外,与营养丰富的生长环境相比,低钾环境更有利于青蒿素的合成。相比之下,适当浓度的硼、砷、镉胁迫可促进青蒿素的积累,而过高的金属元素胁迫则会抑制青蒿素的合成[67]。生物胁迫也能增加青蒿素的产生,大果球囊霉Glomus macrocarpum可增加腺毛密度及青蒿素产量[67]。
JA、ABA和SA在植物应答逆境胁迫过程中发挥着至关重要的作用,是重要的应答非生物胁迫的调控因子。JA和植物受胁迫有关,当植物受盐、干旱、伤及紫外线等胁迫时,植物体内的JA信号增强[54];ABA通过依赖或非依赖的信号传导途径调节渗透压[58];SA参与了植物的干旱胁迫、冷应激、热应激和重金属离子胁迫的应答[68]。JA生物合成相关基因(LOX1、LOX2、AOC和JAR1)受冷胁迫的诱导,冷胁迫通过增加内源性JA提高青蒿素的产量,表明环境胁迫通过激素介导的信号通路影响青蒿素合成及腺毛密度[69]。
5.2 诱导因子对青蒿素合成的影响除以上因素外,内生真菌诱导因子、化学信号诱导因子、糖类、二甲基亚砜(DMSO)、光照等也能影响青蒿素的合成。
5.2.1 内生真菌诱导因子内生真菌共生可作为一种次生代谢诱导剂,促进植物次生代谢产物的产生[70]。研究证明,在青蒿不定根中引入内生真菌炭疽病Colletotrichum sp.[71-72],青蒿素的量提高到13 mg/L。另有报道指出,印度梨形孢(DSM 11827)和固氮菌(W-5)通过增加青蒿植株高度、干质量和叶子生物量实现总生物量的增加,最终影响青蒿素的产量[73]。
5.2.2 化学信号诱导因子NO及ROS作为一种信号应答因子,能促进次生代谢产物的产生。在青蒿素生物合成中,NO和ROS促使DHAA转变成青蒿素,从而使青蒿素量增加[74-75]。
5.2.3 糖类寡糖、寡半乳糖醛酸诱导因子促使根产生大量NO和ROS[74, 76],提高青蒿素的量;葡萄糖[77-78]能促进青蒿素的合成,但果糖能抑制青蒿素的合成。
5.2.4 DMSODMSO是由海洋微生物产生的,其可通过增加根苗中ROS和H2O2浓度,促进青蒿素的合成[79-80]。
5.2.5 光照青蒿素的合成与光合作用有关。将拟南芥中调节光信号的蓝光受体(CRY1)转入青蒿中,青蒿素合成途径关键基因FPS、ADS和CYP71AV1的表达量均增加,青蒿素量也提高30%~40%[81]。此外,还有研究表明,白光能增加青蒿不定根培养过程中青蒿素的形成[82];进一步实验证明,在白光、红光、蓝光、黄光和绿光中,红光更能促进青蒿不定根培养过程中青蒿素的合成[80]。
6 展望青蒿素及其衍生物具有多种生物活性。除能用于对抗疟疾外,青蒿素类药物还能杀灭血吸虫、弓形虫、利什曼原虫等多种寄生虫,治疗红斑狼疮及类风湿性关节炎等免疫性疾病,对多种人源性肿瘤细胞具有较大的杀伤力[83]。虽然青蒿素可通过化学合成、生物半合成[84]、悬浮细胞培养[85]、毛状根培养[86]等方式获得,但青蒿素的主要来源仍然是通过青蒿植株获得。鉴于青蒿原植物中青蒿素的量很低,因此可通过各种代谢调控手段提高青蒿素的量。
随着人们对青蒿素生物合成过程的认识,大量青蒿素相关基因被克隆鉴定,基于过表达生物合成关键基因、抑制支路基因及多基因组合表达的次生代谢工程策略在青蒿素代谢调控中获得长足发展;另外,转录因子、植物激素、逆境及诱导因子对青蒿素积累的研究正在不断深入,不同影响因素对青蒿素合成的分子机制逐步被揭示,这些成果的取得对利用植株获得青蒿素具有重要的指导意义。腺毛是青蒿素生物合成的场所,通过调控腺毛的形态或数量,可从青蒿素合成的特定部位提高青蒿素的产量,从而实现优质品种的培育。
图 2总结了影响青蒿素合成的因素,包括外界环境刺激、激素和非青蒿素生物合成的其他相关基因。其中,外界环境刺激包括逆境(黄色线条中标识的内容)和诱导因子(紫色线条中标识的内容);激素包括JA、GA、SA和ABA;基因主要为已证明与青蒿素产量相关的转录因子及其相关蛋白,另外也包括与腺毛发育相关(黑色标识)和与青蒿素转运相关(黑色标识)的基因。同时,图 2还用颜色区分了激素与基因之间的关系,激素能调控相同颜色标志的基因表达;从左到右,左边的因素能影响右边的因素,最后通过基因影响青蒿素生物合成及腺毛发育。可通过对左边3种因素的调控,开展基于青蒿素生物合成途径的常规次生代谢调控策略和基于腺毛的新型代谢调控策略,提高青蒿素的产量。
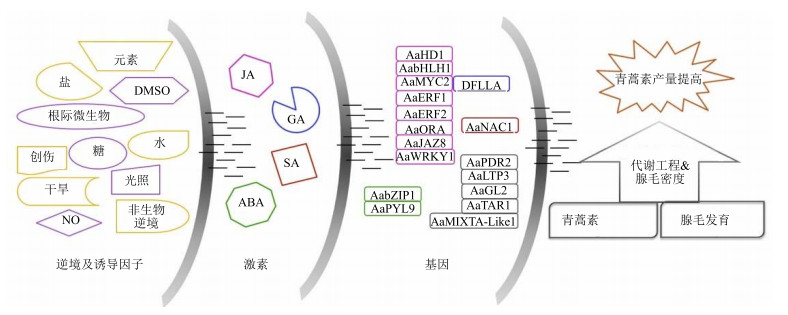
|
图 2 青蒿素代谢工程示意图 Fig.2 Sketch map of artemisinin metabolic engineering |
青蒿素代谢调控可从青蒿素生物合成及支路途径、转录因子、植物激素、逆境、诱导因子和腺毛方面入手,具体包含以青蒿素生物合成途径为基础的常规次生代谢调控和以腺毛为基础的新型代谢调控(图 2)。基于此,本文提出几点新的常规次生代谢工程策略:探究最佳的基因组合表达形式(新的合成途径基因之间及其与支路途径之间的组合),同时结合转运蛋白并应用最适特异启动子;加强多个转录因子联合调控青蒿素产量的研究,全面实现对多个青蒿素生物合成及支路途径的共同调控;联合应用植物激素、逆境及诱导因子,同时调控多个青蒿生物合成相关转录因子和青蒿素生物合成及支路途径的多个不同作用的基因。另一方面,开展基于腺毛密度的新型代谢工程策略:联合调控腺毛发育相关的转录因子、植物激素、逆境及诱导因子,促进多个腺毛发育相关基因的表达,实现青蒿素代谢宏观调控。随着青蒿素生物合成及相关影响因素分子机制的深入研究,多因素青蒿代谢调控得以合理实现,这将为优质高产转基因青蒿品系的培育和青蒿种质遗传改良奠定基础。
| [1] | Pulice G, Pelaz S, Matías-Hernández L. Molecular farming in Artemisia annua, a promising approach to improve anti-malarial drug production[J]. Front Plant Sci, 2016. DOI:10.3389/fpls.2016.00329 |
| [2] | Van Noorden R. Demand for malaria drug soars[J]. Nature, 2010, 466(7307): 672–673. DOI:10.1038/466672a |
| [3] | 张小波, 郭兰萍, 黄璐琦. 我国黄花蒿中青蒿素含量的气候适宜性等级划分[J]. 药学学报, 2011, 46(4):472–478. |
| [4] | Schramek N, Wang H, Römisch-Margl W, et al. Artemisinin biosynthesis in growing plants of Artemisia annua. A 13CO2 study[J]. Phytochemistry, 2010, 71(2/3): 179–187. |
| [5] | Nguyen K T, Arsenault P R, Weathers P J. Trichomes+roots+ROS=artemisinin:regulating artemisinin biosynthesis in Artemisia annua L[J]. In Vitro Cell Dev Pl, 2011, 47(3): 329–338. DOI:10.1007/s11627-011-9343-x |
| [6] | Wang H, Han J, Kanagarajan S, et al. Trichome-specific expression of the amorpha-4, 11-diene 12-hydroxylase (cyp71av1) gene, encoding a key enzyme of artemisinin biosynthesis in Artemisia annua, as reported by a promoter-GUS fusion[J]. Plant Mol Biol, 2013, 81(1/2): 119–138. |
| [7] | Wang Y, Yang K, Jing F, et al. Cloning and characterization of trichome specific promoter of cpr71av1 gene involved in artemisinin biosynthesis in Artemisia annua L.[J]. Mol Biol, 2011, 45(5): 817–824. |
| [8] | Teoh K H, Polichuk D R, Reed D W, et al. Artemisia annua L. (Asteraceae) trichome-specific cDNAs reveal CYP71AV1, a cytochrome P450 with a key role in the biosynthesis of the antimalarial sesquiterpene lactone artemisinin[J]. Febs Lett, 2006, 580(5): 1411–1416. DOI:10.1016/j.febslet.2006.01.065 |
| [9] | Wu T, Wang Y, Guo D. Investigation of glandular trichome proteins in Artemisia annua L. using comparative proteomics[J]. PLoS One, 2012. DOI:10.1371/journal.pone.0041822 |
| [10] | Muangphrom P, Seki H, Fukushima E O, et al. Artemisinin-based antimalarial research:application of biotechnology to the production of artemisinin, its mode of action, and the mechanism of resistance of Plasmodium parasites[J]. J Nat Med, 2016, 70(3): 318–334. DOI:10.1007/s11418-016-1008-y |
| [11] | Suberu J, Gromski P S, Nordon A. Multivariate data analysis and metabolic profiling of artemisinin and related compounds in high yielding varieties of Artemisia annua field-grown in Madagascar[J]. J Pharm Biomed Anal, 2016, 117: 522–531. DOI:10.1016/j.jpba.2015.10.003 |
| [12] | Teoh K H, Polichuk D R, Reed D W, et al. Molecular cloning of an aldehyde dehydrogenase implicated in artemisinin biosynthesis in Artemisia annua[J]. Botany, 2009, 87(6): 635–642. DOI:10.1139/B09-032 |
| [13] | Ryden A M, Carolien R S, Quax W, et al. The molecular cloning of dihydroartemisinic aldehyde reductase and its implication in artemisinin biosynthesis in Artemisia annua[J]. Planta Med, 2010, 76(15): 1778–1783. DOI:10.1055/s-0030-1249930 |
| [14] | Mercke P, Crock J, Croteau R, et al. Cloning, expression, and characterization of epi-cedrol synthase, a sesquiterpene cyclase from Artemisia annua L.[J]. Arch Biochem Biophys, 1999, 369(2): 213–222. DOI:10.1006/abbi.1999.1358 |
| [15] | Cai Y, Jia J W, Crock J, et al. A cDNA clone for[J]. Phytochemistry, 2002, 61(5): 523–529. DOI:10.1016/S0031-9422(02)00265-0 |
| [16] | Bertea C M, Voster A, Verstappen F W A, et al. Isoprenoid biosynthesis in Artemisia annua:cloning and heterologous expression of a germacrene A synthase from a glandular trichome cDNA library[J]. Arch Biochem Biophys, 2006, 448(1/2): 3–12. |
| [17] | Brown G D. The biosynthesis of artemisinin (Qinghaosu) and the phytochemistry of Artemisia annua L. (Qinghao)[J]. Molecules, 2010, 15(11): 7603–7698. DOI:10.3390/molecules15117603 |
| [18] | Picaud S, Brodelius M, Brodelius P E. Expression, purification and characterization of recombinant (E)-[J]. Phytochemistry, 2005, 66(9): 961–967. DOI:10.1016/j.phytochem.2005.03.027 |
| [19] | Zhang L, Jing F, Li F, et al. Development of transgenic Artemisia annua (Chinese wormwood) plants with an enhanced content of artemisinin, an effective anti-malarial drug, by hairpin-RNA-mediated gene silencing[J]. Biotechnol Appl Bioc, 2009, 52(3): 199–207. DOI:10.1042/BA20080068 |
| [20] | Yan L, Ye H C, Wang H, et al. Molecular cloning, Escherichia coli expression and genomic organization of squalene synthase gene from Artemisia annua[J]. Acta Botanica Sinica, 2003, 45(5): 608–613. |
| [21] | Nafis T, Akmal M, Ram M, et al. Enhancement of artemisinin content by constitutive expression of the HMG-CoA reductase gene in high-yielding strain of Artemisia annua L.[J]. Plant Biotechnol Rep, 2011, 5(5): 53–60. |
| [22] | Olsson M E, Olofsson L M, Lindahl A L, et al. Localization of enzymes of artemisinin biosynthesis to the apical cells of glandular secretory trichomes of Artemisia annua L.[J]. Phytochemistry, 2009, 70(9): 1123–1128. DOI:10.1016/j.phytochem.2009.07.009 |
| [23] | Banyai W, Kirdmanee C, Mii M, et al. Overexpression of farnesyl pyrophosphate synthase (FPS) gene affected artemisinin content and growth of Artemisia annua L.[J]. Plant Cell Tiss Org, 2010, 103(2): 255–265. DOI:10.1007/s11240-010-9775-8 |
| [24] | Polichuk D, Teoh K H, Zhang Y, et al. Nucleotide sequence encoding an alcohol dehydrogenease from Artemisia annua and uses thereof:WO, 2010012074[P]. 2011-06-30. |
| [25] | Polichuk D R, Zhan Y S, Reed D W, et al. A glandular trichome-specific monoterpene alcohol dehydrogenase from Artemisia annua[J]. Phytochemistry, 2010, 71(11-12): 1264–1269. DOI:10.1016/j.phytochem.2010.04.026 |
| [26] | Shen Q, Chen Y F, Wang T, et al. Overexpression of the cytochrome P450 monooxygenase (cyp71av1) and cytochrome P450 reductase (cpr) genes increased artemisinin content in Artemisia annua (Asteraceae)[J]. Gen Mol Res, 2012, 11(3): 3298–3309. DOI:10.4238/2012.September.12.13 |
| [27] | Lv Z, Zhang F, Pan Q, et al. Branch pathway blocking in Artemisia annua is a useful method for obtaining high yield artemisinin[J]. Plant Cell Physiol, 2016, 57(3): 588–602. DOI:10.1093/pcp/pcw014 |
| [28] | 王亚雄, 龙世平, 曾利霞, 等. 过表达HDR和ADS基因对青蒿素生物合成的影响[J]. 药学学报, 2014(9):1346–1352. |
| [29] | Tang K, Shen Q, Yan T, et al. Transgenic approach to increase artemisinin content in Artemisia annua L.[J]. Plant Cell Rep, 2014, 33(4): 605–615. DOI:10.1007/s00299-014-1566-y |
| [30] | Duke S O, Paul R N. Development and fine structure of the glandular trichomes of Artemisia annua L.[J]. Int J Plant Sci, 1993, 154: 107–118. DOI:10.1086/297096 |
| [31] | Duke M V, Paul R N, Elsohly H N, et al. Localization of artemisinin and artemisitene in foliar tissues of glanded and glandless biotypes of Artemisia annua L.[J]. Int J Plant Sci, 1994, 155(3): 365–372. DOI:10.1086/297173 |
| [32] | Olofsson L, Lundgren A, Brodelius P E. Trichome isolation with and without fixation using laser microdissection and pressure catapulting followed by RNA amplification:expression of genes of terpene metabolism in apical and sub-apical trichome cells of Artemisia annua L.[J]. Plant Sci, 2012, 183(1): 9–13. |
| [33] | Bryant L, Patole C, Cramer R. Proteomic analysis of the medicinal plant Artemisia annua:data from leaf and trichome extracts[J]. Data Brief, 2016, 7: 325–331. DOI:10.1016/j.dib.2016.02.038 |
| [34] | Xiao L, Tan H, Zhang L. Artemisia annua glandular secretory trichomes:the biofactory of antimalarial agent artemisinin[J]. Sci Bull, 2016, 61(1): 26–36. DOI:10.1007/s11434-015-0980-z |
| [35] | Brown G D, Sy L K. In vivo transformations of artemisinic acid in Artemisia annua plants[J]. Tetrahedron, 2007, 63(18): 9548–9566. |
| [36] | Graham I A, Besser K, Blumer S, et al. The genetic map of Artemisia annua L. identifies loci affecting yield of the antimalarial drug artemisinin[J]. Science, 2010, 327(5963): 328–331. DOI:10.1126/science.1182612 |
| [37] | Arsenault P R, Vail D, Wobbe K K, et al. Reproductive development modulates gene expression and metabolite levels with possible feedback inhibition of artemisinin in Artemisia annua[J]. Plant Physiol, 2010, 154(2): 958–968. DOI:10.1104/pp.110.162552 |
| [38] | Singh N D, Kumar S, Daniell H. Expression of β-glucosidase increases trichome density and artemisinin content in transgenic Artemisia annua plants[J]. Plant Biotechnol J, 2016, 14(3): 1034–1045. DOI:10.1111/pbi.2016.14.issue-3 |
| [39] | Wang B, Kashkooli A B, Sallets A, et al. Transient production of artemisinin in Nicotiana benthamiana, is boosted by a specific lipid transfer protein from A. annua[J]. Metab Eng, 2016, 38: 159–169. DOI:10.1016/j.ymben.2016.07.004 |
| [40] | Han J, Wang H, Kanagarajan S, et al. Promoting artemisinin biosynthesis in Artemisia annua plants by substrate channeling[J]. Mol Plant, 2016, 9(6): 946–948. DOI:10.1016/j.molp.2016.03.004 |
| [41] | Han J, Wang H, Lundgren A, et al. Effects of overexpression of AaWRKY1 on artemisinin biosynthesis in transgenic Artemisia annua plants[J]. Phytochemistry, 2014, 102(6): 89–96. |
| [42] | Ma D, Pu G, Lei C, et al. Isolation and characterization of AaWRKY1, an Artemisia annua transcription factor that regulates the amorpha-4, 11-diene synthase gene, a key gene of artemisinin biosynthesis[J]. Plant Cell Physiol, 2009, 50(12): 2146–2161. DOI:10.1093/pcp/pcp149 |
| [43] | Lv Z, Wang S, Zhang F, et al. Overexpression of a novel NAC domain-containing transcription factor gene (AaNAC1) enhances the content of artemisinin and increases tolerance to drought and Botrytis cinerea in Artemisia annua[J]. Plant Cell Physiol, 2016, 57(9): 1961–1971. DOI:10.1093/pcp/pcw118 |
| [44] | Ji Y, Xiao J, Shen Y, et al. Cloning and characterization of AabHLH1, a bHLH transcription factor that positively regulates artemisinin biosynthesis in Artemisia annua[J]. Plant Cell Physiol, 2014, 55(9): 1592–1604. DOI:10.1093/pcp/pcu090 |
| [45] | Shen Q, Lu X, Yan T, et al. The jasmonate-responsive AaMYC2 transcription factor positively regulates artemisinin biosynthesis in Artemisia annua[J]. New Phytol, 2016, 210(4): 1269–1281. DOI:10.1111/nph.13874 |
| [46] | Shen Q, Cui J, Fu X Q, et al. Cloning and characterization of DELLA genes in Artemisia annua[J]. Genet Mol Res, 2015, 14(3): 10037–10049. DOI:10.4238/2015.August.21.10 |
| [47] | Zhang F, Fu X, Lv Z, et al. A basic leucine zipper transcription factor, AabZIP1, connects abscisic acid signaling with artemisinin biosynthesis in Artemisia annua L.[J]. Mol Plant, 2015, 8(1): 163–175. DOI:10.1016/j.molp.2014.12.004 |
| [48] | Yan T, Chen M, Shen Q, et al. Homeodomain protein 1 is required for jasmonate-mediated glandular trichome initiation in Artemisia annua[J]. New Phytol, 2016. DOI:10.1111/nph.14205 |
| [49] | Yu Z X, Li J X, Yang C Q, et al. The jasmonate-responsive AP2/ERF transcription factors AaERF1 and AaERF2 positively regulate artemisinin biosynthesis in Artemisia annua L.[J]. Mol Plant, 2012, 5(2): 353–365. DOI:10.1093/mp/ssr087 |
| [50] | Lu X, Zhang L, Zhang F, et al. AaORA, a trichome-specific AP2/ERF transcription factor of Artemisia annua, is a positive regulator in the artemisinin biosynthetic pathway and in disease resistance to Botrytis cinerea[J]. New Phytol, 2013, 198(4): 1191–1202. DOI:10.1111/nph.12207 |
| [51] | Tan H, Xiao L, Gao S, et al. Trichome and artemisinin regulator 1 is required for trichome development and artemisinin biosynthesis in Artemisia annua[J]. Mol Plant, 2015, 8(9): 1396–411. DOI:10.1016/j.molp.2015.04.002 |
| [52] | Wang H, Liu W, Qiu F, et al. Molecular cloning and characterization of the promoter of aldehyde dehydrogenase gene from Artemisia annua[J]. Biotechnol Appl Biochem, 2016. DOI:10.1002/bab.1520 |
| [53] | Jindal S, Longchar B, Singh A, et al. Promoters of AaGL2 and AaMIXTA-Like1 genes of Artemisia annua direct reporter gene expression in glandular and non-glandular trichomes[J]. Plant Signal Behav, 2015. DOI:10.1080/15592324.2015.1087629 |
| [54] | Wasternack C. Action of jasmonates in plant stress responses and development-applied aspects[J]. Biotechnol Adv, 2014, 32(1): 31–39. DOI:10.1016/j.biotechadv.2013.09.009 |
| [55] | De Geyter N, Gholami A, Goormachtig S, et al. Transcriptional machineries in jasmonate-elicited plant secondary metabolism[J]. Trends Plant Sci, 2012, 17(6): 349–359. DOI:10.1016/j.tplants.2012.03.001 |
| [56] | Liu S, Tian N, Li J, et al. Isolation and identification of novel genes involved in artemisinin production from flowers of Artemisia annua using suppression subtractive hybridization and metabolite analysis[J]. Planta Med, 2009, 75(14): 1542–1547. DOI:10.1055/s-0029-1185809 |
| [57] | Maes L, Van Nieuwerburgh F C, Zhang Y, et al. Dissection of the phytohormonal regulation of trichome formation and biosynthesis of the antimalarial compound artemisinin in Artemisia annua plants[J]. New Phytol, 2010, 189(1): 176–189. |
| [58] | Yoshida T, Mogami J, Yamaguchi-Shinozaki K. ABA-dependent and ABA-independent signaling in response to osmotic stress in plants[J]. Curr Opin Plant Biol, 2014, 21(21C): 133–139. |
| [59] | Jing F, Zhang L, Li M, et al. Abscisic acid (ABA) treatment increases artemisinin content in Artemisia annua by enhancing the expression of genes in artemisinin biosynthetic pathway[J]. Biologia, 2009, 64(2): 319–323. |
| [60] | Zhang F, Lu X, Lv Z, et al. Overexpression of the Artemisia orthologue of ABA receptor, AaPYL9, enhances ABA sensitivity and improves artemisinin Content in Artemisia annua L.[J]. PLoS One, 2009. DOI:10.1371/journal.pone.0056697 |
| [61] | Pu G, Ma D, Chen J, et al. Salicylic acid activates artemisinin biosynthesis in Artemisia annua L.[J]. Plant Cell Rep, 2009, 28(7): 1127–1135. DOI:10.1007/s00299-009-0713-3 |
| [62] | Aftab T, Khan M M A, Idrees M, et al. Methyl jasmonate counteracts boron toxicity by preventing oxidative stress and regulating antioxidant enzyme activities and artemisinin biosynthesis in Artemisia annua L.[J]. Protoplasma, 2011, 248(3): 601–612. DOI:10.1007/s00709-010-0218-5 |
| [63] | Zhang Y S, Ye H C, Liu B Y, et al. Exogenous GA3 and flowering induce the conversion of artemisinic acid to artemisinin in Artemisia annua plants[J]. Russ J Plant Physl, 2005, 52(1): 58–62. DOI:10.1007/s11183-005-0009-6 |
| [64] | Banerjee S, Zehra M, Gupta M M, et al. Agrobacterium rhizogenes-mediated transformation of Artemisia annua:production of transgenic plants[J]. Planta Med, 1997, 63(5): 467–469. DOI:10.1055/s-2006-957737 |
| [65] | Paniego N, Giulietti A. Artemisinin production by Artemisia annua L. transformed organ cultures[J]. Enzyme Microb Technol, 1996, 18(7): 526–530. DOI:10.1016/0141-0229(95)00216-2 |
| [66] | Robert-Seilaniantz A, Grant M, Jones J D. Hormone crosstalk in plant disease and defense:more than just jasmonate-salicylate antagonism[J]. Annu Rev Phytopathol, 2011, 49(1): 317–343. DOI:10.1146/annurev-phyto-073009-114447 |
| [67] | Pandey N, Pandey-Rai S. Updates on artemisinin:an insight to mode of actions and strategies for enhanced global production[J]. Protoplasma, 2016, 253(1): 15–30. DOI:10.1007/s00709-015-0805-6 |
| [68] | Rivas-San Vicente M, Plasencia J. Salicylic acid beyond defence:its role in plant growth and development[J]. J Exp Bot, 2011, 62(10): 3321–3338. DOI:10.1093/jxb/err031 |
| [69] | Liu W, Wang H, Chen Y, et al. Cold stress improves the production of artemisinin depending on the increase of endogenous jasmonate[J]. Biotechnol Appl Biochem, 2016. DOI:10.1002/bab.1493 |
| [70] | Tan R X, Zou W X. Endophytes:a rich source of functional metabolites[J]. Nat Prod Rep, 2001, 18(4): 448–459. DOI:10.1039/b100918o |
| [71] | Lu H, Zou W X, Meng J C, et al. New bioactive metabolites produced by Colletotrichum sp. an endophytic fungus in Artemisia annua[J]. Plant Sci, 2000, 151(1): 67–73. DOI:10.1016/S0168-9452(99)00199-5 |
| [72] | Wang Y, Zhang H, Zhao B, et al. Improved growth of Artemisia annua L. hairy roots and artemisinin production under red light conditions[J]. Biotechnol Lett, 2001, 23(23): 1971–1973. DOI:10.1023/A:1013786332363 |
| [73] | Arora M, Saxena P, Choudhary D K, et al. Dual symbiosis between Piriformospora indica and Azotobacter chroococcum enhances the artemisinin content in Artemisia annua L.[J]. World J Microbiol Biotechnol, 2016. DOI:10.1007/s11274-015-1972-5 |
| [74] | Zheng L P, Zhang B, Zou T, et al. Nitric oxide interacts with reactive oxygen species to regulate oligosaccharide-induced artemisinin biosynthesis in Artemisia annua hairy roots[J]. J Med Plants Res, 2010, 4(9): 758–766. |
| [75] | Wang J W, Zheng L P, Zhang B, et al. Stimulation of artemisinin synthesis by combined cerebroside and nitric oxide elicitation in Artemisia annua hairy roots[J]. Appl Microbiol Biot, 2009, 85(2): 285–292. DOI:10.1007/s00253-009-2090-9 |
| [76] | Zhang B, Zou T, Lu Y H, et al. Stimulation of artemisinin biosynthesis in Artemisia annua hairy roots by oligogalacturonides[J]. Afr J Biotechnol, 2010, 9(23): 3437–3442. |
| [77] | Weathers P J, DeJesus-Gonzalez L, Kim Y J, et al. Alteration of biomass and artemisinin production in Artemisia annua hairy roots by media sterilization method and sugars[J]. Plant Cell Rep, 2004, 23(6): 414–418. DOI:10.1007/s00299-004-0837-4 |
| [78] | Wang Y. Sugar control of artemisinin production[D]. Massachusetts:Worcester Polytechnic Institute, 2006. |
| [79] | Mannan A, Liu C, Arsenault P R, et al. DMSO triggers the generation of ROS leading to an increase in artemisinin and dihydroartemisinic acid in Artemisia annua shoot cultures[J]. Plant Cell Rep, 2010, 29(2): 143–152. DOI:10.1007/s00299-009-0807-y |
| [80] | Wallaart T E, Pras N, Quax W J. Isolation and identification of dihydroartemisinic acid hydroperoxide from Artemisia annua:a novel biosynthetic precursor of artemisinin[J]. J Nat Prod, 1999, 62(8): 1160–1162. DOI:10.1021/np9900122 |
| [81] | Hong G J, Hu W L, Li J X, et al. Increased accumulation of artemisinin and anthocyanins in Artemisia annua expressing the Arabidopsis blue light receptor CRY1[J]. Plant Mol Biol Rep, 2009, 27(3): 334–341. DOI:10.1007/s11105-008-0088-6 |
| [82] | Liu C, Guo C, Wang Y, et al. Effect of light irradiation on hairy root growth and artemisinin biosynthesis of Artemisia annua L.[J]. Process Biochem, 2002, 38(4): 581–585. DOI:10.1016/S0032-9592(02)00165-6 |
| [83] | 尹纪业, 王和枚, 丁日高. 青蒿素及其衍生物毒理学研究进展[J]. 中国药理学与毒理学杂志, 2014, 28(2):309–314. |
| [84] | 方欣, 卢山, 于宗霞, 等. 青蒿素的生物合成研究[J]. 科技导报, 2015, 33(20):31–35. |
| [85] | Di Sansebastiano G P, Rizzello F, Durante M, et al. Subcellular compartmentalization in protoplasts from Artemisia annua cell cultures:engineering attempts using a modified SNARE protein[J]. J Biotechnol, 2015, 202: 146–152. DOI:10.1016/j.jbiotec.2014.11.016 |
| [86] | Patra N, Srivastava A K. Use of model-based nutrient feeding for improved production of artemisinin by hairy roots of Artemisia annua in a modified stirred tank bioreactor[J]. Appl Biochem Biotechnol, 2015, 177(2): 373–388. DOI:10.1007/s12010-015-1750-8 |
 2017, Vol. 48
2017, Vol. 48


