1994年Jofuku等[1]首次发现了AP2/ERF转录因子在模式植物拟南芥Arabidopsis thaliana (L.) Heynh中调控花发育。1995年Ohmetakagi等[2]分离到的烟草蛋白ERF1、ERF2、ERF3、ERF4与乙烯诱导病程相关。随后,AP2/ERF转录因子的发现与功能研究不断深入。在结构上,AP2/ERF家族成员至少含有1个非常保守的DNA结合区,即AP2/EREBP结合域,该结构域由60个左右氨基酸组成,其N端存在1个碱性亲水区,包含3个反向平行的β折叠,通过β折叠上的精氨酸和色氨酸残基与靶基因双螺旋结构大沟上的8个碱基相连以与GCC-box结合[3]。2002年Sakuma等[4]对拟南芥基因组中全部AP2/ERF进行了序列结构分析,将AP2/ERF类转录因子分为5个亚族:AP2、ERF、RAV、CBF/DREB和特异蛋白AL079349即Soloist。CBF/DREB和ERF亚族(共121个成员)具有1 个AP2结构域和1个保守的WLG基序;AP2亚族(17个成员)具有2个AP2结构域;RAV亚族(6个成员)除具有AP2结构域外还含有1个B3结构;AL079349蛋白没有典型的WLG基序。前期研究认为AP2/ERF是植物中特有的一类转录因子,但Wessler[5]和Magnani等[6]相继在嗜热四膜虫Tetrahymena thermophila、束毛藻Trichodesmium erythraeum、病毒(噬菌体x01和噬菌体RB49)中也发现了编码AP2/ERF的基因[7]。2013年,有研究表明在细菌、动物中也存在与AP2/ERF同源性较高的基因[8]。Magnani等[6]认为植物中的AP2/ERF基因很有可能起源于细菌或者病毒基因的横向转移。
近20年的研究表明,AP2/ERF转录因子参与调节多种植物生理功能,包括植物生长[9, 10]、花发育[11]、果实发育[12]、种子发育[13],以及植物对损伤、病菌防御、盐、干旱等胁迫应答[14, 15, 16, 17, 18, 19, 20, 21, 22, 23];另外还参与水杨酸、茉莉酸、乙烯、脱落酸等信号转导途径[21, 24, 25],是逆境信号交叉途径中的连接因子[26]。AP2/ERF是植物中最大的转录因子家族之一。目前在重要农作物中已发现超过1 065个AP2/ERF类转录因子。拟南芥、水稻Oryza sativa L.、玉米Zea mays L. 和大豆Glycine max (L.) Merr. 基因组中分别含有166、188、330和381个AP2/ERF类转录因子[27]。
药用植物是医药健康领域发展的重要物质基础,但一些重要植物药的资源短缺,给患者用药安全带来一定隐患[28]。植物药的主要活性成分通常是药用植物的次生代谢产物,通过代谢工程调控生产药用植物次生代谢产物,在解决资源短缺、创制新药方面具有广阔前景。药用植物次生代谢产物的合成与调控研究已越来越引起人们的关注。合成生物学的兴起更掀起了药用植物代谢途径和调控研究的热潮。在代谢途径酶基因克隆和功能分析、利用酵母等宿主合成药用植物代谢产物方面均已取得显著进展[29, 30]。转录调控是植物代谢调控的重要方面,已发现多种转录因子如WRKY、bZIP、bHLH及AP2/ERF等均参与植物萜类、黄酮类、生物碱类代谢合成的调控[3, 31]。植物对逆境胁迫的反应往往与植物次生代谢产物合成相关[30, 32],水分胁迫诱导干旱相关的代谢反应,由于气孔关闭、CO2的摄入显著降低,导致通过卡尔文循环固定CO2所需的还原当量(NADPH+H+)的消耗显著下降,因而形成较大的氧胁迫和还原当量的过剩。结果,代谢过程转向消耗还原当量的那些物质的生物合成相应的被还原的天然产物,如异戊二烯类、酚类或生物碱类化合物的生物合成增强[29]。由此参与植物逆境胁迫反应的转录因子存在调控植物次生代谢产物合成的可能。Ramakrishna等[33]综述了转录因子在植物次生代谢调控中的重要性及转录因子与植物萜类积累的相关性。本文重点综述AP2/ERF类转录因子在调控药用植物主要活性成分生物合成中的作用。
1 AP2/ERF对青蒿素合成的调控青蒿素(artemisinin)是从药用植物黄花蒿中提取的有过氧基团的倍半萜内酯抗疟药。青蒿素是中国发现的第一个被国际公认的天然药物,具有毒性低、抗疟性强等特点,被WTO批准为世界范围内治疗脑型疟疾和恶性疟疾的首选药物。青蒿素的生物合成途径经过几十年的研究逐渐清晰。如图 1所示,首先通过甲羟戊酸(MVA)途径和2-C-甲基-D-赤藓糖醇-4-磷酸(MEP)途径[34]生成异戊烯基焦磷酸(IPP)和二甲基烯丙基焦磷酸(DMAPP);其次在法尼基焦磷酸合酶(FPPS)的催化下合成法尼基焦磷酸(FPP);然后在紫穗槐-4,11-二烯合酶(ADS)的催化下合成紫穗槐-4,11-二烯;接着由紫穗槐-4,11-二烯氧化酶(AMO)催化合成青蒿醇和青蒿醛;青蒿醛受青蒿醛双键还原酶2(DBR2)或青蒿醛双键还原酶1(DBR1)催化而还原成双氢青蒿醛;再由青蒿醇或者双氢青蒿醛经氧化反应生成青蒿酸或者二氢青蒿酸;最后经过一系列的酶促反应或者其他反应合成青蒿素[35]。
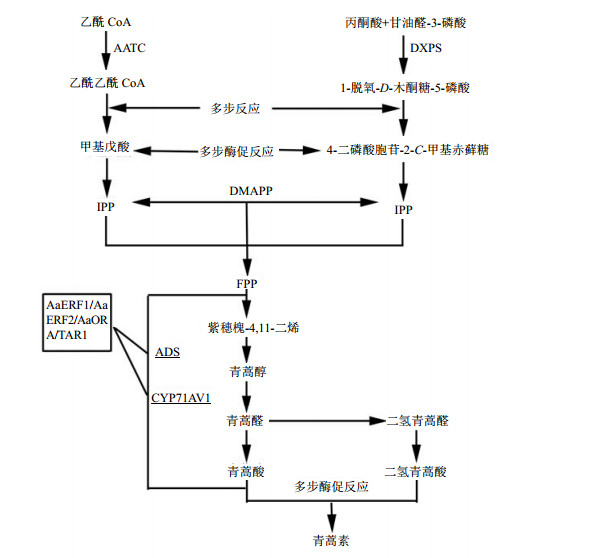 |
加下划线字体部分为AP2/ERF类转录因子主要调控的酶基因,方框内为转录因子,下同;AATC-乙酰辅酶A酰基转移酶 DXPS-1-脱氧-D-木酮糖-5-磷酸合酶 IPP-异戊烯基焦磷酸 DMAPP-二甲基烯丙基焦磷酸 FPP-法尼基焦磷酸 ADS-紫穗槐-4,11-二烯合酶 AMS-紫穗槐- 4,11-二烯氧化酶 DBR1/DBR2-青蒿醛双键还原酶 CYP71AV1-紫穗槐二烯P450氧化酶 A1DH1-二氢青蒿醛脱氢酶 The underlined part is the enzyme gene that AP2/ERF transcription factor major regulation,the transcription factor is in the box,same as below; AATC- acetyl coenzyme A acyltransferase DXPS-1-deoxy-D-xylulose-5-phosphate synthase IPP-isopentenyl pyrophosphate DMAPP-dimethylallyl pyrophosphate FPP-farnesyl pyrophosphate ADS-amorpha-4,11-diene synthase AMS-amorpha-4,11-diene oxidase DBR1/DBR2-double artesunate acid reductase CYP71AV1-amorphadiene P450 oxidase A1DH1-dihydro-artesunate aldehyde dehydrogenase 图 1 青蒿素生物合成途径 Fig.1 Biosynthetic pathway of artemisinin |
有研究利用酵母单杂交技术分离出了2个茉莉酸(JA)响应的AP2家族转录因子AaERF1和AaERF2[3, 36]。它们能够与存在于ADS和紫穗槐二烯P450氧化酶(CYP71AV1)启动子中的倍半萜合酶(CBF2)和RAA元件结合,并与ADS和CYP71AV1基因协同表达。CYP71AV1参与催化合成青蒿素前体紫穗槐-4,11-二烯到青蒿酸的一系列过程,是青蒿素生物合成途径中的关键酶。AaERF1和AaERF2在烟草中瞬时表达可增强ADS和CYP71AV1的启动子活性,并且过量表达转基因青蒿植物表明ADS和CYP71AV1的转录水平均有所提高,青蒿素和青蒿酸的积累也增加。与此相反,在沉默AaERF1或AaERF2的转基因品系中青蒿素合成被抑制。这些结果表明,AaERF1和AaERF2是青蒿素的生物合成的2个正向调控因子。
通过对从青蒿中克隆的6个AP2类转录因子的组织特异性表达分析筛选出1个在表皮毛特异表达的转录因子AaORA[37]。AaORA的表达模式与ADS、CYP71AV1和DBR2相似。通过转基因实验对AaORA基因过表达或抑制,可明显上调或下调青蒿中ADS、CYP71AV1、DBR2的表达水平,表明AaORA转录因子也能够正向调控青蒿素和青蒿酸的合成。
2015年Tan等[38]用相似的方法从青蒿中分离出调控毛状体和青蒿素生物合成途径的AP2/ERF转录因子TAR1。TAR1过表达和抑制表达的青蒿转基因株系中青蒿素的量明显的相应上调和下降。实验表明TAR1很可能直接作用于ADS和CYP71AV1,从而正向调控青蒿素生物合成。另外有研究表明,脱落酸(ABA)和JA处理可显著增加青蒿素量[39, 40, 41],而转录因子ERF3在乙烯或JA诱导下转录水平均有所提高。推测ERF3与青蒿素的积累也有一定的关系[42]。
2 AP2/ERF对紫杉醇合成的调控紫杉醇(taxol)为抗癌药,每年总市值逾10亿元[43]。1962年从短叶红豆杉Taxus brevifolia Nutt. 树皮中首次分离到紫杉醇,已被批准用于治疗乳腺癌和肺癌[44]。其需求量大,野生资源濒危,市用紫杉醇主要来源于化学半合成。紫杉醇合成途径中,牻牛儿基牻牛儿基焦磷酸(GGPP)合成之前的步骤与青蒿素合成途径类似(图 2)。GGPP作为前体物质在紫杉烯合酶(TASY)的催化下生成紫杉烯。紫杉烯又经一系列酶催化反应合成紫杉烯化合物的A、B、C环,再经多步紫杉烷羟基化酶连续氧化其中包括紫杉烯5α-羟基化酶、紫杉烯醇5α-乙酰氧化基转移酶(T5αH)、紫杉烷10β-羟基化酶(T10βH)、紫杉烷7β-羟基化酶、紫杉烷2α-羟基化酶、紫杉烷13α-羟基化酶(T13αH)、紫杉烷2α-苯甲基酰基转移酶、紫杉烷14β-羟基化酶、C13-苯基丙酸-侧链-CoA转移酶、紫杉烷C13-侧链-N-苯甲酰转移酶、10-去乙酰巴卡亭III-10β-乙酰转移酶等,最终生成紫杉醇[45]。
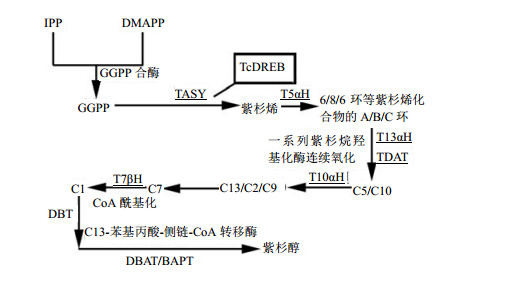 |
GGPP-牻牛儿基牻牛儿基焦磷酸 TASY-紫杉烯合酶 T5αH-5α-乙酰氧化基转移酶 T13αH-紫杉烷13α-羟基化酶 TDAT-紫杉烷酰基转移酶T10αH-紫杉烷-10β-羟基化酶 T7βH-紫杉烷-7β-羟基化酶 DBT-2α-O-苯甲酰转移酶 BAPT-苯基丙酸-CoA转移酶 DBAT-10-去乙酰巴卡亭III-10β-乙酰转移酶 GGPP-geranyl geranyl pyrophosphate TASY-taxadiene synthase T5αH-5α-acetoxy-transferase T13αH-taxane13α-hydroxylase TDAT-taxane acyltransferase T10αH-taxane-10β-hydroxylase T7βH-taxane7β-hydroxylase DBT-2α-O-benzoyl transferase BAPT-phenylpropionic acid transferase-CoA DBAT-10-deacetyl baccatin III-10β-acetyltransferase 图 2 紫杉醇生物合成途径 Fig.2 Biosynthetic pathway of taxol |
2009年Dai等[46]通过JA处理东北红豆杉悬浮细胞发现紫杉醇的量升高。利用酵母单杂实验从中分离出与AP2/ERF家族其他转录因子具有较高同源性的TcAP2,表明TcAP2可能调节胁迫诱导的靶基因开关,通过调控紫杉醇生物合成途径中关键酶基因的表达而增加紫杉醇的积累。Dai等[46]又发现了东北红豆杉中另一个AP2类转录因子TcDREB。TcDREB能够与紫杉醇的合成途径中的TASY、T10βH、T13αH和T5αH的启动子及TASY基因的启动子区中茉莉酸甲酯(MeJA)响应元件GCC-Box结合,可见TcDREB转录因子可能参与东北红豆杉异戊二烯代谢途径产物紫杉醇的合成[47]。
3 AP2/ERF对吲哚萜类生物碱合成的调控长春花Catharanthus roseus (L.) G. Don是夹竹桃科长春花属的一种多年生草本植物[48]。其乳汁中含有多种生物碱,如长春花碱和长春新碱,作为多种癌症如白血病、哈杰金氏症的治疗药物,在临床上得到了广泛应用[49]。文多灵和长春质碱的调血脂作用也非常显著。目前长春花中萜类吲哚生物碱(TIAs)的生物合成途径也基本清晰(图 3)。合成途径上游阶段首先通过吲哚途径多步酶促反应生成色胺和萜类途径生成裂环马钱子苷;色胺和裂环马钱子苷经异胡豆苷合成酶(STR)催化耦合成异胡豆苷。异胡豆苷是长春花TIAs途径的共同前体物质。合成途径下游阶段异胡豆苷经过3个支路多步催化反应分别形成文多灵、长春质碱、蛇根碱,最终在血红素类氧化酶多酶促反应下形成长春碱或长春新碱[50, 51]。
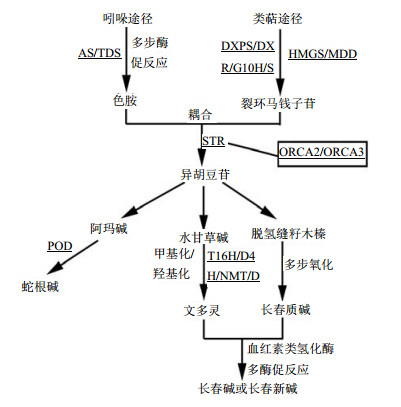 |
AS-苯甲酸合成酶 TDS-色氨酸脱羧酶 DXR-5-磷酸脱氧木酮糖还原酶 G10H-牻牛儿醇-10羟化酶 DXPS-5-磷酸脱氧木酮糖合酶 SLS-马钱子苷合成酶 HMGS-3-羟基-3-甲基戊二酸单酰辅酶A合酶 MDD-甲羟戊酸5-焦磷酸脱羧酶 STR-异胡豆苷合成酶 POD-过氧化酶 T16H-它波宁羟化酶 D4H-去乙酰文多灵-4-脱羧酶 NMT-N-甲基转移酶 DAT-脱乙酰文多灵-4-O-乙酰转移酶 AS-benzoic acid synthase TDS-tryptophan decarboxylase DXR-deoxy- 5-phosphate wood xylulose reductase G10H-geraniol-10-hydroxylase DXPS-deoxyxylulose5-phosphate synthase SLS-strychnos synthase HMGS-3-hydroxy-3-methyl-glutaryl coenzyme A synthetase MDD-5- mevalonate pyrophosphate decarboxylase STR-strictosidine synthase POD-peroxidase T16H-tabersonine-16-hydroxylase D4H-deacetylation vindoline-4-decarboxylase NMT-N-methyltransferase DAT-deacetylation vindoline -4-O-acetyltransferase 图 3 长春碱生物合成途径 Fig.3 Biosynthetic pathway of vinblastine |
1999年Menke等[52]用酵母单杂交方法从长春花中筛选出2个AP2/ERF类转录因子ORCA1和ORCA2,随后将其转入长春花悬浮细胞,结果表明ORCA2可以使STR 42 bp区域包含1个GCC-Box元件,被激活而提高酶基因的转录水平,首次发现GCC-Box和ORCA2在JA诱导响应的萜类吲哚生物碱的生物合成中具有重要作用。van de Fits等[53]在马达加斯加长春花Catharanthus roseus (L.) Don中使用了转移DNA(T-DNA)激活标签法,分离出ORCA3基因。过量表达ORCA3,长春碱生物合成途径中的色氨酸脱羧酶(TDC)、STR、异胡豆苷- β-D-葡萄糖苷酶(SGD)、细胞色素P450还原酶(CPR)、去乙酰文多灵-4-脱羧酶(D4H)等关键酶基因表达量均提高,另外色胺和色氨酸的量也有所提高。表明其可以诱导萜类吲哚前体的生成。EMSA和粒子轰击瞬时表达实验表明,ORCA3直接作用于TDC、STR、CPR转录激活的DNA结合蛋白。ORCA3主要受MeJA[54]诱导,且JA生物合成途径的前体和中间体的合成也会诱发ORCA3的表达,主要是通过与引发子反应原件(JERE)直接作用于STR基因表达[55]。张鑫等[56]和Zhou等[57]用MeJA和硝普钠分别处理长春花毛状根,进一步证实ORCA3转录水平可作为长春碱积累的重要信号。Wang等[58]将香叶醇10羟化酶(G10H)和ORCA3共转化或G10H独立转化到长春花根中,发现长春碱的量明显上调。大量研究[59, 60, 61, 62]表明,ORCA3在调控长春碱生物合成过程中具有重要作用。
4 AP2/ERF对烟草烟碱合成的调控烟碱(nicotine)是茄科植物中的一种杂环化合物,属于生物碱类物质,也是N胆碱受体激动药的代表,可用于N1和N2受体及神经系统,也是人类吸食烟草的主要成分。烟草为我国主要经济作物。烟草烟碱又称尼古丁。烟碱的生物合成是由前体物质鸟氨酸或者精氨酸经脱羧酶催化生成腐胺开始。在腐胺N-甲基转移酶(PMT)的催化下生成N-甲基腐胺,再经N-甲基腐胺氧化酶(MPO)的催化生成4-甲基丁醛。4-甲基丁醛和天冬氨酸经吡啶核苷酸循环途径生成烟酸,再经过反应生成的吡啶烷经自然环化生成吡咯烷,最后缩合形成烟碱(图 4)[63]。
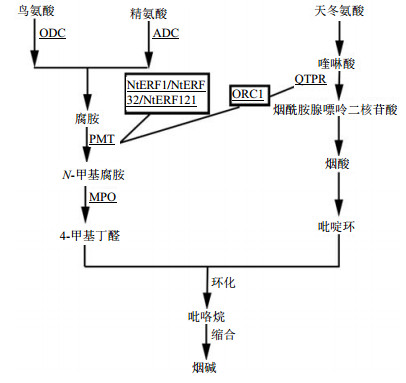 |
ODC-鸟氨酸脱羧酶 ADC-精氨酸脱羧酶 QTPR-喹啉酸核糖转移酶 PMT-腐胺 N-甲基转移酶 MPO-N-甲基腐胺氧化酶 ODC-ornithine decarboxylase ADC-arginine decarboxylase QTPR- quinolinic acid ribose PMT-putrescine methyl N-transferase MPO-N- methyl putrescine oxidase 图 4 烟草烟碱合成途径 Fig.4 Biosynthetic pathway of tobacco nicotine |
2011年De Boer等[64]研究发现,从长春花中分离的AP2/ERF类转录因子ORC1正调控几个结构基因编码参与烟碱生物合成的酶,在培养的烟草根中过表达ORC1可刺激烟碱的合成。而且,ORC1需要启动子中同时含有CCC基序和GCC-Box才能最大程度促进烟碱生物合成,主要是通过少量JA诱导,正调控烟碱生物合成基因PMT和QPRT[65]。实验证明NIC是编码烟草烟碱生物合成途径中的7个基因组成的基因簇。用JA处理烟草,检测到NtERF1、NtERF32、NtERF121的表达量增加。NtPMT1a是编码烟碱吡咯烷环形成的腐胺N-甲基转移酶的重要基因。实验表明NtERF32异位过表达可以使NtPMT1a转录水平提高,总生物碱和烟碱的量提高。因此,这些AP2/ERF转录因子是烟草烟碱生物合成途径中的重要转录因子[66]。
5 AP2/ERF对其他活性成分合成的调控枇杷Eriobotrya japonica (Thunb.) Lindl. 果实中已分离出18个AP2/ERF类基因。其中EjAP2-1的表达与水果木质化呈负相关。双荧光素酶分析表明,EjAP2-1可以抑制拟南芥和枇杷木质素生物合成基因的启动子的活性。酵母双杂交和双分子荧光互补测定表明,EjAP2-1和木质素相关的EjWYB1或者EjWYB2存在蛋白互作。此外,EjAP2-1和EjWYB1或者EjWYB2组合对启动子Ej4CL1具有抑制作用。同时ERF基序突变的EjAP2-1(mEjAP2-1)仍可与EjWYB蛋白互作,但失去了这种抑制作用。因此,EjAP2-1是木质素生物合成的间接转录阻遏[67]。
紫草素(alkannin)是一种结晶性粉末,存在于紫草科植物紫草Lithospermum erythrorhizon Sieb. et Zucc. 根中,具有抗癌、抗炎、抗菌等作用。研究发现白、蓝、红光条件下,紫草中LeERF-1表达量显著下调,但红光相对于其他光线条件的抑制作用比较薄弱。组织特异性表达表明,LeERF-1在黑暗生长的土壤根部表达。这些模式均与不同光信号调节紫草素及其衍生物影响一致,表明LeERF-1可能对紫草素的生物合成起正向调节作用[68]。
6 结语在利用大规模测序挖掘酶基因和调控基因研究迅速发展起来之前,就有学者指出转录因子是研究通过生物工程生产植物代谢活性成分的重要工具[69, 70]。利用转录因子调控代谢途径的优势在于有些转录因子不仅能调控途径中的单个酶基因,往往可以调控多个酶基因协同表达,可有效启动和关闭次生代谢合成途径,从而调节特定次生代谢物的合成。还可以改变代谢产物产生的植物组织器官,甚至在天然不产生某种代谢产物的植物中调控产生新的代谢产物。从第一个AP2/ERF转录因子发现至今,有关其作用的研究已取得较大进展。但在药用植物中鉴定的调控代谢产物生物合成的AP2/ERF类转录调控因子还较少。AP2/ERF类转录调控因子在其他农作物和经济作物中也可见调控代谢产物合成的相关报道,如Li等[71]已鉴定EREB58调控玉米倍半萜烯合成;在苹果中已发现AP2/ERF类转录因子可调控类胡萝卜素生物合成途径中关键酶的表达[72];葡萄中也发现AP2/ERF类转录因子参与果实成熟过程ACC氧化酶、萜类合酶和脂氧合酶的基因表达调控[73]。AP2/ERF转录因子还参与植物地上部分表皮蜡质生物合成的调控[74]。但AP2/ERF调控药用植物及其他作物代谢产物生物合成方面的研究仍有待进一步深入。AP2/ERF调控特异靶基因的机制及不同调控因子和信号系统间的相互作用还不是很清楚,还需进一步研究。
| [1] | Jofuku K D, Denboer B G W, Vanmontagu M, et al. Control of arabidopsis flower and seed development by the homeotic gene APETALA2[J]. Plant Cell, 1994, 6(9):1211-1225. |
| [2] | Takagi O, Shinshi H. Ethylene-inducible DNA binding proteins that interact with an ethylene-responsive element[J]. Plant Cell, 1995, 7(2):173-182. |
| [3] | 季爱加, 罗红梅, 徐志超, 等. 药用植物转录因AP2/ERF研究与展望[J]. 科学通报, 2015, 60(14):1272-1284. |
| [4] | Sakuma Y, Liu Q, Dubouzet J G, et al. DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration-and cold-inducible gene expression[J]. Biochem Biophys Res Commun, 2002, 290(3):998-1009. |
| [5] | Wessler S R. Homing into the origin of the AP2 DNA binding domain[J]. Trends Plant Sci, 2005, 10(2):54-56. |
| [6] | Magnani E, Sjölander K, Hake S. From endonucleases to transcription factors:evolution of the AP2 DNA binding domain in plants[J]. Plant Cell, 2004, 16(9):2265-2277. |
| [7] | 秦捷, 王武, 左开井, 等. AP2基因家族的起源和棉花AP2转录因子在抗病中的作用[J]. 棉花学报, 2005, 17(6):366-370. |
| [8] | Kazuhiko Y, Takanori K, Motoaki S, et al. DNA-binding domains of plant-specific transcription factors:structure, function, and evolution[J]. Trends Plant Sci, 2013, 18(5):267-276. |
| [9] | Gao C, Li P, Song A, et al. Isolation and characterization of six AP2/ERF transcription factor genes in chrysanthemum nankingense[J]. Int J Mol Sci, 2015, 16(1):2052-2065. |
| [10] | Lee S Y, Hwang E Y, Seok H Y, et al. Arabidopsis AtERF71/HRE2 functions as transcriptional activator via cis-acting GCC box or DRE/CRT element and is involved in root development through regulation of root cell expansion[J]. Plant Cell Rep, 2015, 34(2):223-231. |
| [11] | Huala E, Sussex I M. LEAFY interacts with floral homeotic genes to regulate Arabidopsis floral development[J]. Plant Cell, 1992, 4(8):901-913. |
| [12] | Chung M Y, Vrebalov J, Alba R, et al. A tomato (Solanumly copersicum) APETALA2/ERF gene, SlAP2a, is a negative regulator of fruit ripening[J]. Plant J, 2010, 64(6):936-947. |
| [13] | Niu X, Helentjaris T, Bate N J. Maize ABI4 binds coupling element1 in abscisic acid and sugar response genes[J]. Plant Cell, 2002, 14(10):2565-2575. |
| [14] | Djemal R, Khoudi H. Isolation and molecular characterization of a novel WIN1/SHN1 ethylene- responsive transcription factor TdSHN1 from durum wheat (Triticumturgidum L. subsp. durum)[J]. Protoplasma, 2015, 252(6):1-13. |
| [15] | Lin R C, Park H J, Wang H Y. Role of Arabidopsis RAP2. 4 in regulating light- and ethylene-mediated develop- mental processes and drought stress tolerance[J]. Mol Plant, 2008, 1(1):42-57. |
| [16] | Liu N, Zhong N Q, Wang G L, et al. Cloning and functional characterization of PpDBF1 gene encoding a DRE-binding transcription factor from Physcomitrella patens[J]. Planta, 2007, 226(4):827-838. |
| [17] | Qin F, Kakimoto M, Sakuma Y, et al. Regulation and functional analysis of ZmDREB2A in response to drought and heat stresses in Zea mays L.[J]. Plant J, 2007, 50(1):54-69. |
| [18] | Wang X, Chen X, Liu Y, et al. CkDREB gene in Caraganakorshinskii is involved in the regulation of stress response to multiple abiotic stresses as an AP2/EREBP transcription factor[J]. Mol Biol Rep, 2011, 38(4):2801-2811. |
| [19] | Yamaguchi-Shinozaki K, Shinozaki K. Characterization of the expression of a desiccation-responsive rd29 gene of Arabidopsis thaliana and analysis of its promoter in transgenic plants[J]. Mol Gen Genet, 1993, 236(2/3):331-340. |
| [20] | Zhang G, Chen M, Chen X, et al. Phylogeny, gene structures, and expression patterns of the ERF gene family in soybean (Glycine max L.)[J]. J Exp Bot, 2008, 59(15):4095-4107. |
| [21] | Kim Y H, Jeong J C, Park S, et al. Molecular characterization of two ethylene response factor genes in sweetpotato that respond to stress and activate the expression of defense genes in tobacco leaves[J]. J Plant Physiol, 2012, 169(11):1112-1120. |
| [22] | Wang X, Han H, Yan J, et al. A new AP2/ERF transcription factor from the oil plant Jatropha curcasconfers salt and drought tolerance to transgenic tobacco[J]. Appl Biochem Biotechnol, 2015, 176(2):582-597. |
| [23] | Dong L, Cheng Y, Wu J, et al. Overexpression of GmERF5, a new member of the soybean EAR motif-containing ERF transcription factor, enhances resistance to phytophthorasojae in soybean[J]. J Exp Bot, 2015, 66(9):2635-2647. |
| [24] | Craig S, Ling Y. Regulation of specialized metabolism by WRKY transcription factors[J]. Plant Physiol, 2015, 167(2):295-306. |
| [25] | Moore M, Vogel M O, Dietz K J. The acclimation response to high light is initiated within seconds as indicated by upregulation of AP2/ERF transcription factor network in Arabidopsis thaliana[J]. Plant Signal Behav, 2014, 9(10):976479. |
| [26] | Gao J S, Hu L, Xie P, et al. Isolation and molecular characterization of an ethylene response[J]. J Biosci, 2014, 39(5):887-897. |
| [27] | 张计育, 王庆菊, 郭忠仁. 植物AP2/ERF类转录因子研究进展[J]. 遗传, 2012, 34(7):835-847. |
| [28] | Girardi C L, Rombaldi C V, Dal Cero J, et al. Genome-wide analysis of the AP2/ERF superfamily in apple and transcriptional evidence of ERF involvement in scab pathogenesis[J]. Sci Hortic, 2013, 151(2):112-121. |
| [29] | Chabner B A. Drug shortages-a critical challenge for the generic-drug market[J]. New England J Med, 2011, 365(23):2147-2149. |
| [30] | Dai Z B, Liu Y, Guo J, et al. Yeast synthetic biology for high-value metabolites[J]. FEMS Yeast Res, 2015, 15(1):1-11. |
| [31] | Jensen M K, Keasling J D. Recent applications of synthetic biology tools for yeast metabolic engineering[J]. FEMS Yeast Res, 2015, 15(1):1-10. |
| [32] | Selmar D, Kleinwachter M. Stress enhances the synthesis of secondary plant products:theimpact of stress-related over-reduction on the accumulationof natural products[J]. Plant Cell Physiol, 2013, 54(6):817-826. |
| [33] | Ramakrishna A, Ravishankar G. Influence of abiotic stress signals on secondary metabolites in plants[J]. Plant Signal Behav, 2011, 6(11):1720-1731. |
| [34] | Laule O, Fürholz A, Chang H S, et al. Crosstalk between cytosolic and plastidial pathways of isoprenoid biosynthesis in Arabidopsis thaliana[J]. Proc Nat Acad Sci USA, 2003, 100(11):6866-6871. |
| [35] | 孔建强, 王伟, 程克棣, 等. 青蒿素的合成生物学研究进展[J]. 药学学报, 2013, 48(2):193-205. |
| [36] | Lu X, Jiang W, Zhang L, et al. Characterization of a novel ERF transcription factor in Artemisia annua and its induction kinetics after hormones and stress treatments[J]. Mol Biol Rep, 2012, 39(10):9521-9527. |
| [37] | Lu X, Zhang L, Zhang F, et al. AaORA, a trichome-specific AP2/ERF transcription factor of Artemisia annua, is a positive regulator in the artemisinin biosynthetic pathway and in disease resistance to Botrytis cinerea[J]. New Phytol, 2013, 198(4):1191-1202. |
| [38] | Tan H, Xiao L, Gao S, et al. Trichome and artemisinin regulator 1 is required for trichomedevelopment and artemisinin biosynthesis in Artemisia annua L.[J]. Mol Plant, 2015, 8(9):1396-1411. |
| [39] | Guo X X, Yang X Q, Yang R Y, et al. Salicylic acid and methyl jasmonate but not Rose Bengal enhance artemisinin production through invoking burst of endogenous singlet oxygen[J]. Plant Sci, 2010, 178(4):390-397. |
| [40] | Lu X, Lin X, Shen Q, et al. Characterization of the jasmonate biosynthetic gene allene oxide cyclase in Artemisia annua L., source of the antimalarial drug artemisinin[J]. Plant Mol Biol Rep, 2011, 29(2):489-497. |
| [41] | Lu X, Zhang F, Jiang W, et al. Characterization of the first specific jasmonate biosynthetic pathway gene allene oxide synthase from Artemisia annua[J]. Mol Biol Rep, 2012, 39(3):2267-2274. |
| [42] | Lu X, Jiang W, Zhang L, et al. Characterization of a novel ERF transcription factor in Artemisia annua and its induction kinetics after hormones and stress treatments[J]. Mol Biol Rep, 2012, 39(10):9521-9527. |
| [43] | Malik S, Cusidó R M, Mirjalili M H, et al. Production of the anticancer drug taxol in Taxus baccata suspension cultures:a review[J]. Proc Biochem, 2011, 46(1):23-34. |
| [44] | Howat S, Park B, Oh I S, et al. Paclitaxel:biosynthesis, production and future prospects[J]. New Biotechnol, 2014, 31(3):242-245. |
| [45] | 孔建强, 王伟, 朱平, 等. 紫杉醇生物合成的研究进展[J]. 药学学报, 2007, 42(4):358-365. |
| [46] | Dai Y, Qin Q, Dai D, et al. Isolation and characterization of a novel cDNA encoding methyl jasmonate-responsive transcription factor TcAP2 from Taxus cuspidata[J]. Biotechnol Lett, 2009, 31(11):1801-1809. |
| [47] | 戴怡龄, 唐克轩, 杨金水. 红豆杉中与异戊二烯代谢途径相关的AP2类转录调控因子的克隆与功能研究[D]. 上海:复旦大学, 2008. |
| [48] | Verpoorte R, van der Heijden R, van Gulik. Alkaloids[M]. San Diego:Academic Press, 1991. |
| [49] | 习利平, 宋新波, 张丽娟. 长春碱类抗肿瘤药的研究进展[J]. 药物评价研究, 2011, 34(1):59-62. |
| [50] | 邢世海, 王荃, 潘琪芳, 等. 长春花萜类吲哚生物碱的生物合成途径[J]. 西北植物学报, 2012, 32(9):1917-1927. |
| [51] | 祝冬青, 吴晓俊, 郑珍贵, 等. 长春花生物碱生物合成的分子机理[J]. 国外医药:植物药分册, 2004, 19(3):93-99. |
| [52] | Menke F L H, Champion A, Kijne J W, et al. A novel jasmonate-and elicitor-responsive element in the periwinkle secondary metabolite biosynthetic gene Str interacts with a jasmonate-and elicitor-inducible AP2-domain transcription factor, ORCA2[J]. EMBO J, 1999, 18(16):4455-4463. |
| [53] | van de Fits L, Memelink J. The jasmonate-inducible AP2/ERF-domain transcription factor ORCA3 activates gene expression via interaction with a jasmonate- responsive promoter element[J]. Plant J, 2001, 25(1):43-53. |
| [54] | Goklany S, Rizvi N F, Loring R H, et al. Jasmonate-dependent alkaloid biosynthesis in Catharanthus roseus hairy root cultures is correlated with the relative expression of Orca and Zct transcription factors[J]. Biotechnol Prog, 2013, 29(6):1367-1376. |
| [55] | Courdavault V, Burlat V, St-Pierre B, et al. Proteins prenylated by type I protein geranylgeranyltransferase act positively on the jasmonatesignalling pathway triggering the biosynthesis of monoterpene indole alkaloids in Catharanthus roseus[J]. Plant Cell Rep, 2009, 28(1):83-93. |
| [56] | 张鑫, 宋经元, 胡鸢雷, 等. bHLH转录因子调控植物活性成分生物合成的研究进展[J]. 药学学报, 2014, 49(4):435-442. |
| [57] | Zhou M L, Zhu X M, Shao J R, et al. Transcriptional response of the catharanthine biosynthesis pathway to methyl jasmonate/nitric oxide elicitation in Catharanthusroseus hairy root culture[J]. Applied Microbiol Biotechnol, 2010, 88(3):737-750. |
| [58] | Wang C T, Liu H, Gao X S, et al. Overexpression of G10H and ORCA3 in the hairy roots of Catharanthus roseus improves catharanthine production[J]. Plant Cell Rep, 2010, 29(8):887-894. |
| [59] | Pan Q, Wang Q, Yuan F, et al. Overexpression of ORCA3 and G10H in Catharanthus roseus plants regulated alkaloid biosynthesis and metabolism revealed by NMR-metabolomics[J]. PLoS One, 2012, 7(8):e43038. |
| [60] | van der Fits L, Memelink J. ORCA3, a jasmonate-responsive transcriptional regulator of plant primary and secondary metabolism[J]. Science, 2000, 289(5477):295-297. |
| [61] | Peebles C A M, Hughes E H, Shanks J V, et al. Transcriptional response of the terpenoid indole alkaloid pathway to the overexpression of ORCA3 along with jasmonic acid elicitation of Catharanthus roseus hairy roots over time[J]. Metab Eng, 2009, 11(2):76-86. |
| [62] | Zhou M L, Zhu X M, Shao J R, et al. Transcriptional response of the catharanthine biosynthesis pathway to methyl jasmonate/nitric oxide elicitation in Catharanthus roseus hairy root culture[J]. Appl Microbiol Biotechnol, 2010, 88(3):737-750. |
| [63] | 戚元成, 刘卫群. 烟碱生物合成分子机制的研究进展[J]. 中国烟草学报, 2011(1):87-94. |
| [64] | De Boer K, Tilleman S, Pauwels L, et al. Apetala2/ethylene response factor and basic helix-loop-helix tobacco transcription factors cooperatively mediate jasmonate-elicited nicotine biosynthesis[J]. Plant J, 2011, 66(6):1053-1065. |
| [65] | Sutter V, Vanderhaeghen R, Tilleman S, et al. Exploration of jasmonatesignalling via automated and standardized transient expression assays in tobacco cells[J]. Plant J, 2005, 44(6):1065-1076. |
| [66] | Sears M T, Zhang H, Rushton P J, et al. NtERF32:a non-NIC2 locus AP2/ERF transcription factor required in jasmonate-inducible nicotine biosynthesis in tobacco[J]. Plant Mol Biol, 2014, 84(1/2):49-66. |
| [67] | Zeng J, Li X, Xu Q, et al. EjAP2-1, an AP2/ERF gene, is a novel regulator of fruit lignification induced by chilling injury, via interaction with EjMYB transcription factors[J]. Plant Biotechnol J, 2015, 13(9):1325-1334. |
| [68] | Zhang W, Zou A, Miao J, et al. LeERF-1, a novel AP2/ERF family gene within the B3 subcluster, is down-regulated by light signals in Lithospermum erythrorhizon[J]. Plant Biol, 2011, 13(2):343-348. |
| [69] | Gantet P, Memelink J. Transcription factors:tools to engineer the production of pharmacologically active plant metabolites[J]. Trends Pharmacol Sci, 2002, 23(12):563-569. |
| [70] | Broun P. Transcription factors as tools for metabolic engineering in plants[J]. Curr Opin Plant Biol, 2004, 7(2):202-209. |
| [71] | Li S, Wang H, Li F et al. The maize transcription factor EREB58 mediates jasmonate-induced production of sesquiterpene volatiles[J]. Plant J, 2015, 84(2):296-308. |
| [72] | Charles A D, Nicky D, David L, et al. The Phytoene synthase gene family of apple (Malus x domestica) and its role in controlling fruit carotenoid content[J]. BMC Plant Biol, 2015, 15:185. |
| [73] | Grant R C, Ryan G, Karen A S. Transcriptomic analysis of the late stages of grapevine (Vitis vinifera cv. cabernet Sauvignon) berry ripening reveals significant induction of ethylene signaling and flavor pathways in the skin[J]. BMC Plant Biol, 2014, 14:370. |
| [74] | Young S G, Hyojin K, Hae J K, et al. Arabidopsis cuticular wax biosynthesis is negatively regulated by the DEWAX gene encoding an AP2/ERF-type transcription factor[J]. Plant Cell, 2014, 4(26):1666-1680. |
 2016, Vol. 47
2016, Vol. 47


