二氢乳清酸脱氢酶(dihydroorate dehydrogenase,DHODH)是一种含铁的黄素依赖的线粒体酶,是核酸中嘧啶合成的关键酶,催化从头嘧啶生物合成途径中的第四步反应。抑制二氢乳清酸脱氢酶,可以阻断新生嘧啶合成,致使DNA合成障碍,从而抑制活化淋巴细胞的增殖和B淋巴细胞中的T细胞依赖性自体抗体的形成[1-2]。目前二氢乳清酸脱氢酶已被证实为治疗类风湿关节炎以及其他免疫性疾病的有效靶点[3-4],特立氟胺[5-6]以及阿托伐醌[7]已作为二氢乳清酸脱氢酶抑制剂进行研究,已经上市的来氟米特(leflunomkle,LEF)是临床常用的DHODH有效抑制剂,通过阻断嘧啶的从头合成途径和抑制酪氨酸激酶的活性,阻断细胞信号传导过程,是具有抗炎及免疫抑制作用的异恶唑类衍生物[8]。但由于长期服用来氟米特会导致秃头[9]、肺动脉高压[10]、肾损伤、皮疹[11]、腹泻、肝酶异常以及肺结节形成等不良反应[12-13],寻求新型的、不良反应少的免疫抑制剂成为一项长期工作。
三维定量构效关系(three-dimensional quantitativestructure-activity relationshi,3D-QSAR)研究被广泛应用于各类药物的研究与开发中[14-15]。本研究采用3D-QSAR中常用的比较分子力场分析(CoMFA)和比较分子相似性指数分析(CoMSIA)方法建立模型[16]。通过验证,建立的CoMFA和CoMSIA模型是稳定且具有良好预测能力的。根据三维等势图分析,全面直观了解噻唑类衍生物结构的二氢乳清酸脱氢酶抑制活性。
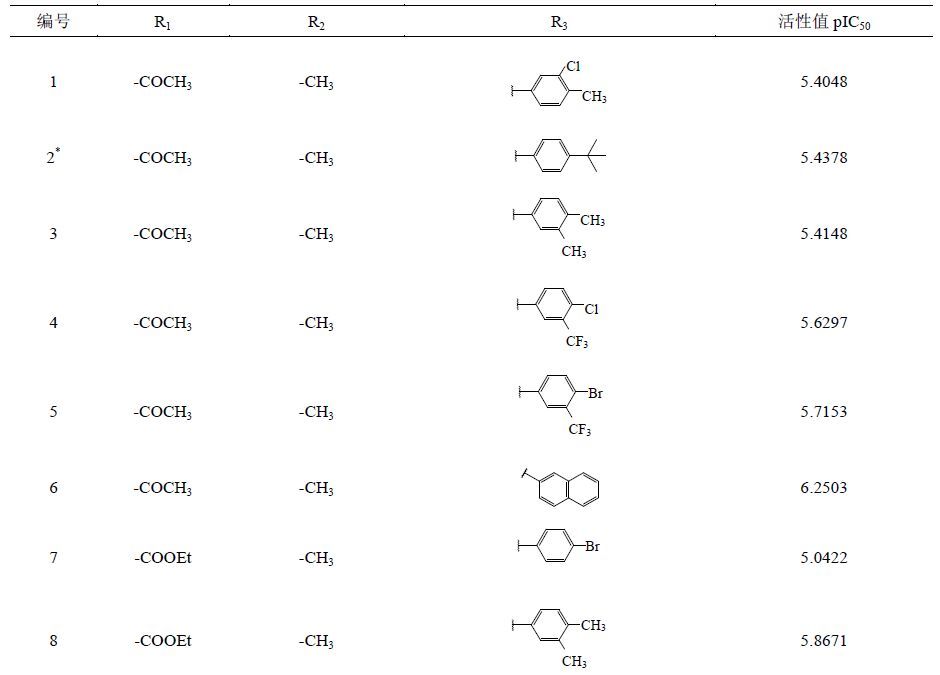
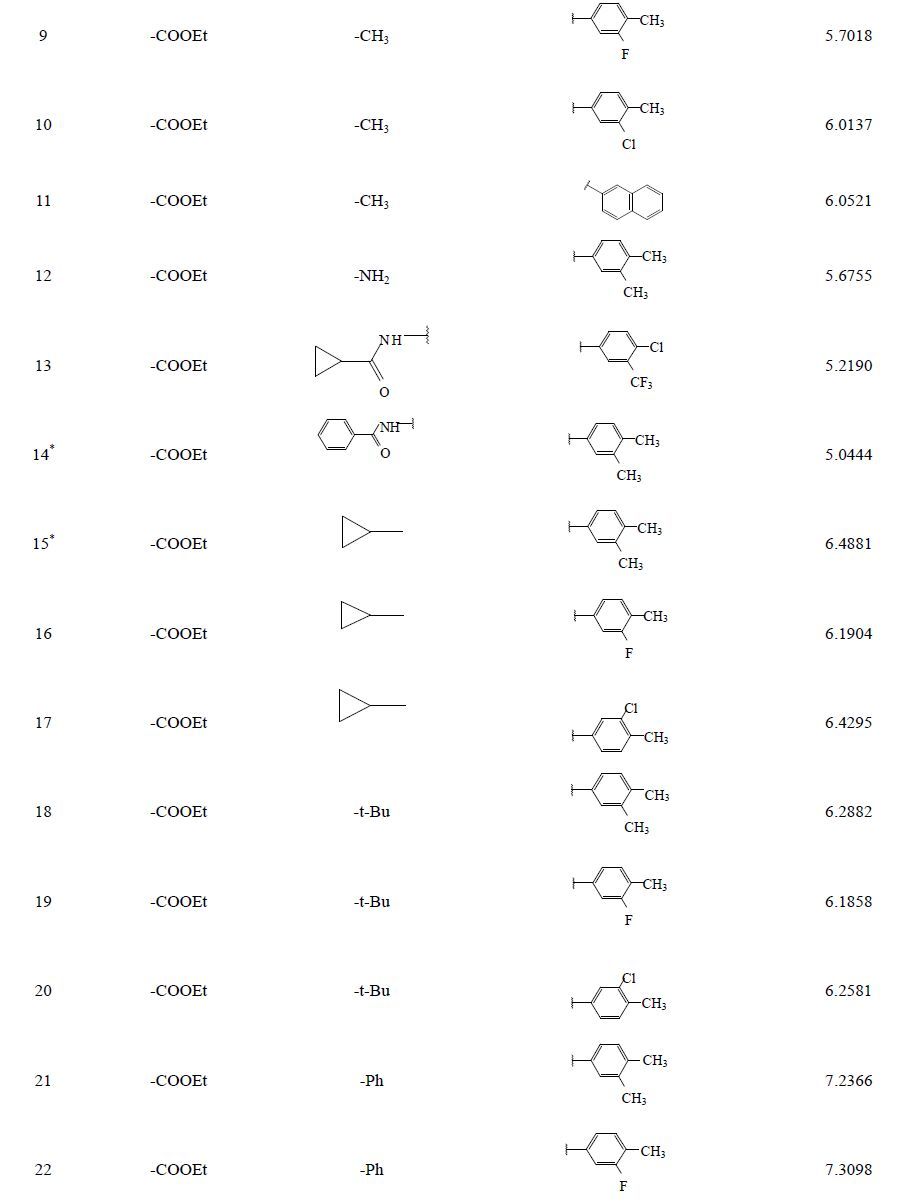
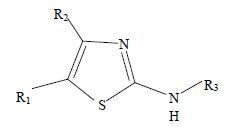
1 材料与方法 1.1 数据优化与叠合噻唑类二氢乳清酸脱氢酶抑制剂的结构与活性来自于参考文献[17]。为了使建立的CoMFA和CoMSIA模型具有可靠性,建模所需的化合物分子必须经过筛选,共筛选出38个化合物分子,其基本骨架见图 1和表 1,根据随机和活性梯度分布原则,化合物分为2组,其中32个化合物作为训练集,6个化合物作为测试集。所有化合物对二氢乳清酸脱氢酶抑制作用均以IC50值表示,IC50值均转换成pIC50值。
|
图 1 噻唑类衍生物基本骨架图 Fig. 1 Structure of thiazole derivatives |
| 表 1 分子结构与抑制二氢乳清酸脱氢酶活性 Table 1 Structure and activity inhibition of DHODH |
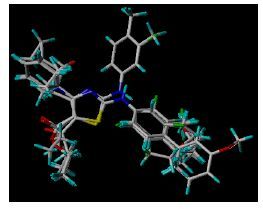
采用Sybyl 2.1搭建,通过在Tripos立场下加Gasteiger-MMFF94电荷对所有分子进行能量优化,其余设置参数均使用默认值。选取最高活性分子35号作为模板,应用Align Database模块将所有优化后的最低能量构象进行叠合,结果见图 2。
|
图 2 分子叠合图 Fig. 2 Superposition of compounds |
1.2 3D-QSAR建模
CoMFA是构建3D-QSAR模型中的一种,特点是配体分子用立体场和静电场来描述并用三维交叉格点取样建模[18-19]。CoMSIA是配体分子用5个场,即立体场、静电场、疏水性场、氢键供体场、氢键受体场,来进行描述[20-21]。在CoMFA建模中,使用偏最小二乘(PLS)进行相互验证方法得到交叉验证系数q2以及最佳主成分数n。最终用非交叉验证方法得到相关系数r2,标准偏差SEE、方差分析F值、立体场S%、静电场E%、疏水场H%、氢键供体场D%、氢键受体场A%等值。CoMSIA模型的建立与CoMFA相似。
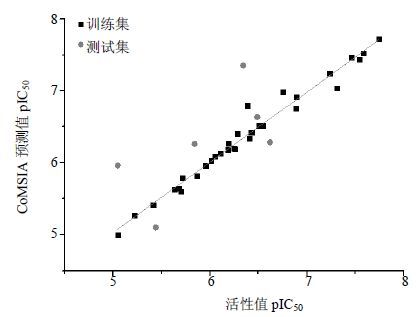
2 结果 2.1 3D-QSAR模型CoMFA和CoMSIA模型总结在表 2中,CoMFA模型包括立体场与静电场,相互验证系数q2=0.796,最佳主成分数n=8,回归系数r2=0.978,标准偏差SEE=0.124。CoMSIA模型包括5个场,相互验证系数q2=0.721,最佳主成分数n=9,回归系数r2=0.976,标准偏差SEE=0.132。这些数值预示了CoMFA和CoMSIA是可信的。表 3中显示了预测值与活性值pIC50。
| 表 2 训练集CoMFA与CoMSIA模型的统计参数 Table 2 Statistical parameters of CoMFA and CoMSIA models for training set model |
| 表 3 化合物实验的活性值(pIC50)与预测值(pIC50) Table 3 Experimental activities (pIC50 value) and predicted activities(pIC50 value) |
2.2 模型验证
通过线性回归,图 3中显示的CoMFA和CoMSIA模型具有很强的预测能力,几乎所有化合物都位于或接近趋势线,说明这两种模型具有较强说服力。

|
图 3 3D-QSAR模型中训练集和测试集预测值数据与活性值数据之间的相关性 Fig. 3 Correlation between experimental and predicted values of 3D-QSAR model for training and test sets |
2.3 CoMFA和CoMSIA模型三维等势图分析
以化合物35为参照样本的CoMFA模型三维等势分析如图 4。绿色表示增大基团会增加化合物活性,黄色表示增大基团会降低化合物活性。图中,可明显看见氨基上的R3取代基有黄色的等势域,也有绿色的等势域。预示着与氨基直接相连的取代基若增大,会减少化合物的活性,如,38号分子R3苯环上有3个取代基团,相较于35号分子,38号R3苯环上多取代2个F,使得黄色区域的空间结构增大,故38号分子活性降低。相反,在R1、R2取代基相同的化合物分子中比较,绿色区域取代基结构较大的4、5、6号分子活性较1、2、3号分子活性大;10、11号分子活性较7、8、9号分子活性大。这与化合物本身的pIC50值一致,表现出很好的分子模型预测。
|
图 4 CoMFA立体静电场三维等势图 Fig. 4 Contuor map of CoMFA steric and electrostatic fields |
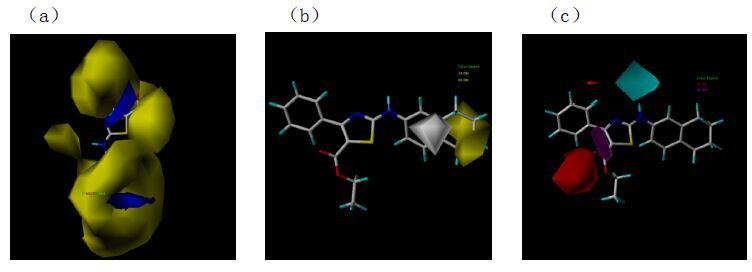
图 5为以35号化合物分子为参照样本的CoMSIA模型三维等势图。其中5a为立体静电场等势图,各个颜色代表的意义与CoMFA模型相同。通过观察比较,CoMSIA模型与CoMFA模型基本保持一致。图 5b为疏水场等势域,黄色等势域代表疏水有利区域,白色等势域代表亲水有利区域。图 5b中氨基所连接的苯环上有大块白色等势域,在此区域增加疏水性有利于活性的增加;而苯环上取代基区域显示大片黄色区域,增加亲水性基团可以提高化合物活性。图 5c为氢键供体与受体的等势分布图。蓝绿色表示增加氢键供体有利于化合物活性,紫色区域表示增加氢键供体不利于化合物活性,而红色区域表示增加氢键受体不利于化合物活性。如,23与26号分子相比较,23号分子R3苯环上多一个甲基,增加了分子的疏水性,故活性大于26号分子;26与27号分子相比较,26号分子R3苯环上连接氯原子,而27号分子连接的是三氟甲基,26号分子R3苯环极性大于27号分子R3苯环,亲水性增强,故26号分子活性较27号分子大。因此根据各个不同的区域,合理选择氢键供体与受体的改造方式,可以优化化合物结构,提高化合物的活性。
|
图 5 CoMSIA立体静电场(a)、疏水场(b)、氢键供体场与受体场(c)三维等势图 Fig. 5 Contuor map of COMSIA steric and electrostatic (a),hydrophobic (b),and H-bonding donor and acceptor (c) fields |
3 讨论
本研究依据38个噻唑类二氢乳清酸脱氢酶抑制剂的结构与活性,用CoMFA和CoMSIA得到稳定且具有较好预测能力的3D-QSAR模型,通过分子对接与计算打分,预测潜在活性分子,并且模拟出对接的三维立体图,显示良好的对接空间结构。通过三维等势图分析可了解其更多的化学结构与生物活性关系,为其提供结构改造的方向。该模型的建立
对二氢乳清酸脱氢酶抑制剂活性和选择性的优化改造具有一定意义,为今后该类化合物设计提供新思路。
| [1] | Shih K C, Lee C C, Tsai C N, et al. Development of a human dihydroorotate dehydrogenase (hDHODH) pharma-similarity index approach with scaffold-hopping strategy for the design of novel potential inhibitors[J]. Plos One, 2014, 9(2):87960. doi:10.1371/journal.pone.0087960 |
| [2] | Marianne L H, Hélène M L, Farah E M, Malmquist N A, et al. Original 2-(3-Alkoxy-1H-pyrazol-1-yl) azines inhibitors of human dihydroorotate dehydrogenase (DHODH)[J]. J MED CHEM, 2015, 58(14):5579–5598. doi:10.1021/acs.jmedchem.5b00606 |
| [3] | Song Y, Zhang Y, Lee A R, et al. Comparison of two molecular scaffolds, 5-methylisoxazole-3-carboxamide and 5-methylisoxazole-4-carboxamide[J]. Curr Pharm Design, 2014, 20(1):146–152. doi:10.2174/13816128113199990584 |
| [4] | Raissi H, Mollania F. Immunosuppressive agent leflunomide:A SWNTs-immobilized dihydroortate dehydrogenase inhibitory effect and computational study of its adsorption properties on zigzag single walled (6,0) carbon and boron nitride nanotubes as controlled drug delivery devices[J]. Eur J Pharm Sci, 2014, 56(22):37–54. |
| [5] | Gerschenfeld G, Servy A, Valeyrieallanore L, et al. Fatal toxic epidermal necrolysis in a patient on teriflunomide treatment for relapsing multiple sclerosis[J]. Mult Scler, 2015, 21(11):2600–2610. |
| [6] | Miller A E. Teriflunomide:a once-daily oral medication for the treatment of relapsing forms of multiple sclerosis[J]. Clin Ther, 2015, 37(10):2366–2380. doi:10.1016/j.clinthera.2015.08.003 |
| [7] | Guler J L, White J 3rd, Phillips M A, et al. Atovaquone tolerance in plasmodium falciparum parasites selected for high-level resistance to a dihydroorotate dehydrogenase inhibitor[J]. Antimicrob Agents Ch, 2015, 59(1):686–689. doi:10.1128/AAC.02347-14 |
| [8] | Elsetouhy D A, Abdelmalak N S, Anis S E, et al. Leflunomide biodegradable microspheres intended for intra-articular administration:development, anti-inflammatory activity and histopathological studies[J]. Int J Pharm, 2015, 495(2):664–670. doi:10.1016/j.ijpharm.2015.09.040 |
| [9] | Lazzarini R, Capareli G C, Buense R, et al. Alopecia universalis during treatment with leflunomide and adalimumab-case report[J]. An Bras Dermatol, 2014, 89(2):416–420. |
| [10] | Alvarez P A, Saad A K, Flagel S, et al. Leflunomide-induced pulmonary hypertension in a young woman with rheumatoid arthritis:a case report[J]. Cardiovasc Toxicol, 2012, 12(2):180–183. doi:10.1007/s12012-012-9153-3 |
| [11] | Pinto B, Dhir V, Krishnan S, et al. Leflunomide-induced DRESS syndrome with renal involvement and vasculitis[J]. Clin Rheumatol, 2013, 32(5):689–693. doi:10.1007/s10067-012-2152-8 |
| [12] | Yoshikawa G T, Dias G A, Fujihara S, et al. Formation of multiple pulmonary nodules during treatment with leflunomide[J]. J Bras Pneumol, 2015, 41(3):281–284. doi:10.1590/S1806-37132015000004247 |
| [13] | Bilasy S E, Essawy S S, Mandour M F, et al. Myelosuppressive and hepatotoxic potential of leflunomide and methotrexate combination in a rat model of rheumatoid arthritis[J]. Pharmacol Rep, 2015, 67(1):102–114. doi:10.1016/j.pharep.2014.08.009 |
| [14] | Zaheer-Ul H, Uddin R, Yuan H, et al. Receptor-based modeling and 3D-QSAR for a quantitative production of the butyrylcholinesterase inhibitors based on genetic algorithm[J]. J Chem Inf Model, 2008, 48(5):1092–1103. doi:10.1021/ci8000056 |
| [15] | Almerico A M, Tutone M, Lauria A. Receptor-guided 3D-QSAR approach for the discovery of c-kit tyrosine kinase inhibitors[J]. J Mol Model, 2012, 18(7):2885–2895. doi:10.1007/s00894-011-1304-0 |
| [16] | Kubinyi H, Folkers G, Martin Y C. 3D-QSAR in drug design[M]. Kluwer Academic, 1998. |
| [17] | Zhu J, Han L, Diao Y, et al. Design, synthesis, X-ray crystallographic analysis, and biological evaluation of thiazole derivatives as potent and selective inhibitors of human dihydroorotate dehydrogenase[J]. J Med Chem, 2015, 58(3):1123–1139. doi:10.1021/jm501127s |
| [18] | Cramer R D, Patterson D E, Bunce J D. Comparative molecular field analysis (CoMFA). 1. Effect of shape on binding of steroids to carrier proteins[J]. J Am Chem Soc, 1988, 110(18):5959-5967. |
| [19] | Clark M, Iii R D C, Jones D M, et al. Comparative molecular field analysis (CoMFA). 2. Toward its use with 3D-structural databases[J]. Tetrahed Comp Meth, 1990, 3(1):47-59. |
| [20] | Klebe G, Abraham U, Mietzner T. Molecular similarity indices in a comparative analysis (CoMSIA) of drug molecules to correlate and predict their biological activity[J]. J Med Chem, 1994, 37(24):4130–4146. doi:10.1021/jm00050a010 |
| [21] | Klebe G, Abraham U. Comparative Molecular Similarity Index Analysis (CoMSIA) to study hydrogen-bonding properties and to score combinatorial libraries[J]. J Comput Aid Mol Des, 1999, 13(1):1–10. doi:10.1023/A:1008047919606 |
 2017, Vol. 40
2017, Vol. 40